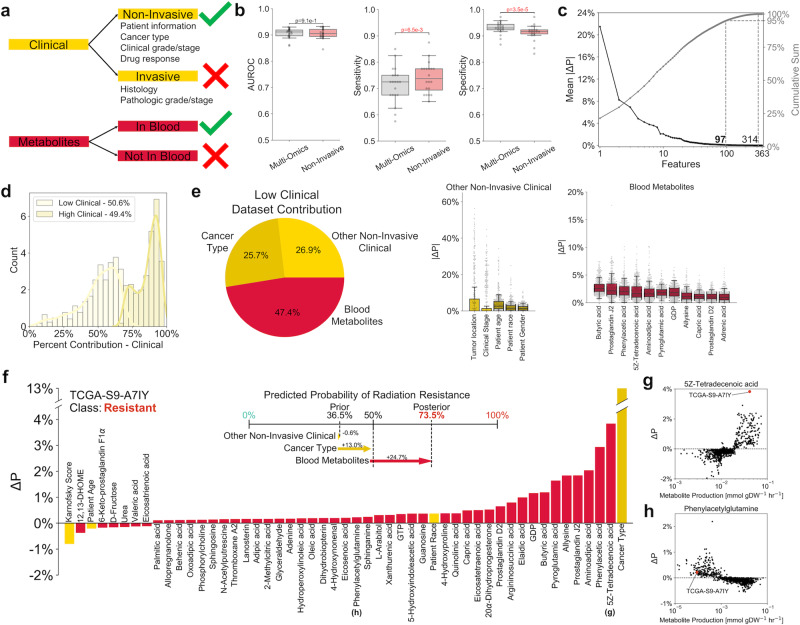
Fig. 6. Non-invasive classifier integrating non-invasive clinical and blood-based metabolite features for prediction of radiation response.
a Schematic showing inclusion and exclusion criteria for features in the non-invasive classifier. b Comparison of model performance between multi-omics and non-invasive classifiers. n = 20 training+validation/testing splits. Boxplots: box = 25th, 50th, and 75th percentiles, whiskers = 1.5 times the interquartile range. Statistical test: two-sided t-test. c (Left, black) Mean absolute SHAP values (mean |ΔP|) for individual features. (Right, gray) Cumulative mean |ΔP| scores. d k-means clustering of samples into “Low” and “High” clinical groups based on the relative contribution of the clinical dataset (Supplementary Fig. 13b). e Clinical and metabolic dataset contributions among the “Low Clinical” group. Individual features with mean |ΔP| scores above 1% are shown. n = 457 samples. Boxplots: box = 25th, 50th, and 75th percentiles, whiskers = 1.5 times the interquartile range. f Breakdown of individual feature contributions toward prediction of radiation response in a representative radiation-resistant TCGA patient (TCGA-S9-A7IY). (Upper) Contribution of each dataset toward the progression from prior to posterior probability of radiation resistance. (Lower) SHAP values for this individual patient. g, h Plots of SHAP value versus predicted metabolite production rate for two metabolic features, illustrating g a feature with large individual importance score relative to other patients (significant utility as a personalized blood-based biomarker), and h a feature with small individual importance score relative to other patients (little utility as a personalized blood-based biomarker).