Abstract
G protein-coupled receptors (GPCRs) comprise the most important superfamily of protein targets in current ligand discovery and drug development. GPCRs are integral membrane proteins that play key roles in various cellular signaling processes. Therefore, GPCR signaling pathways are closely associated with numerous diseases, including cancer and several neurological, immunological, and hematological disorders. Computer-aided drug design (CADD) can expedite the process of GPCR drug discovery and potentially reduce the actual cost of research and development. Increasing knowledge of biological structures, as well as improvements on computer power and algorithms, have led to unprecedented use of CADD for the discovery of novel GPCR modulators. Similarly, machine learning approaches are now widely applied in various fields of drug target research. This review briefly summarizes the application of rising CADD methodologies, as well as novel machine learning techniques, in GPCR structural studies and bioligand discovery in the past few years. Recent novel computational strategies and feasible workflows are updated, and representative cases addressing challenging issues on olfactory receptors, biased agonism, and drug-induced cardiotoxic effects are highlighted to provide insights into future GPCR drug discovery.
Keywords: G protein-coupled receptors, GPCR activation, computer-aided drug design, molecular dynamics, machine learning, structure-based drug design, ligand-based drug design
Impact statement
This review briefly describes current progress in computer-aided drug design and machine learning approaches for the structural study of G-protein-coupled receptors and GPCR-targeted drug discovery. The ever-increasing knowledge of GPCR structure–activity relationships, along with the continuous improvement of computer functions and algorithms, including novel artificial-intelligence-based methodologies, has accelerated drug discovery through the utilization of remarkable computer-aided drug design methods that are continuously improved and updated. New technological improvements in simulations of molecular dynamics have provided further helpful tools for deciphering the dynamic processes and challenges underlying how GPCR structures transduce physiological signals into diverse cellular responses. This is complementary to X-ray crystallography and cryo-electron microscopy (EM), which can be used to determine the three-dimensional finely tuned conformational changes in the active states of a diversity of GPCRs. The further decoding of allosteric/authentic and other molecular mechanisms of specific GPCRs, together with the addition of structure-based and ligand-based drug design that results in rapid collection of three-dimensional structures and the evolution of database technologies, will lead to a deeper understanding of the complexity of specific GPCR-ligand pairs and the development of a powerful platform for GPCR drug discovery. Given that GPCRs represent the important drug targets for current medicine, this review has more general implications and is thus of interest to the broader research and industry communities, including structure- and ligand-based drug design, structural information technology, medicinal chemistry, and drug discovery.
Introduction
G protein-coupled receptors (GPCRs) have been the most widely studied drug targets over the past few decades. This protein superfamily consists of more than 800 protein members, making it the largest superfamily of proteins in the human body.1,2 GPCRs are integral membrane proteins containing seven transmembrane α-helices and are coupled to heterotrimeric G proteins on the intracellular side of the membrane. They are categorized into six classes based on sequence similarities, and classes A, B, C, and F have been discovered in humans. Among them, class A (rhodopsin-like) contains the majority of GPCRs with 719 members.3 Endogenous ligands for GPCRs include from small molecules like lipids, ions, to peptides and proteins.4 In this regard, GPCRs with unknown endogenous ligands are defined as orphan receptors.5 There are approximately 100 endoGPCRs that are characterized as orphan receptors so far.6
GPCRs play essential roles in numerous physiological and pathological mechanisms. They normally respond to a wide variety of endogenous signals and undergo conformational changes that are subsequently transmitted to the G protein or other small proteins like arrestin to cause activation and further transduction of cellular signals. GPCR proteins are involved in various physiological functions, such as cell migration, proliferation, and metabolism, making them vital targets for therapeutic treatment for cancer, HIV infection, and other major GPCR-related diseases.7 Understanding the molecular mechanism of GPCRs has therefore become one of the core issues in drug research and development. GPCRs have historically been of great interest to the pharmaceutical industry, and nearly 40% of the Food and Drug Administration (FDA)-approved drugs are developed for GPCRs and act on more than 100 unique GPCR targets, accounting for around 25% of potentially druggable human GPCRs.7–9 Therefore, it is pertinent to underline that there are still varieties of potentially druggable GPCRs, which are undoubtedly a huge appeal to future discovery of therapeutic agents. Over the past five years, GPCR modulators have continued to hit the market. At this time of writing, 41 drugs targeting GPCRs have been approved by the FDA, while over 142 compounds targeting 83 different GPCRs are currently under clinical trials.3
Fantastic progress has been made in resolving GPCR three-dimensional structures by X-ray crystallography, coupled with lipidic mesophases and cryo-electron microscopy (cryo-EM) since 2007.10–12 Structures of more than 60 unique GPCRs have been determined with over 370 diverse conformational states.3 All available GPCR crystal and cryo-EM structures have now been surveyed and incorporated into an interactive resource integrated as a browsable database called GPCRdb, which provides detailed structural information and appropriate experimental conditions.1,13 Moreover, around 40 additional online databases and servers are available, which focus on the structural information of GPCRs, relevant receptor-ligand interactions, receptor-intracellular partner interactions, and their oligomerization.14 In particular, ice-breaking progress has been made over the past 10 years on a few high-impact structures including agonist-bound receptors, GPCR-G protein complexes, and elusive chemokine receptors, with the advent of new techniques such as protein-based nanodiscs and fusion protein engineering.15–22 Multitudes of three-dimensional GPCRs structures, including active, inactive, and ligand-binding states are disclosed, revealing the general basis for receptor activation, signaling, and allosteric modulation processes. Besides, much effort has been made in recent years towards identifying orphan and adhesion GPCRs, but progress has been limited, making the discovery of novel ligands for these receptors particularly appealing.23
In-depth understanding of the molecular mechanisms for GPCRs, coupled with the abundance of crystal and cryo-EM structures that have come available in the last decades, allows researchers to use computational methodologies in order to explore detailed information in ligand-GPCR complexes (Figure 1).11,13,17,24–27 Computer-aided drug design (CADD) has long been applied as an effective way to expedite the process of GPCR drug discovery and potentially reduce the actual cost of research and development.28 CADD methods generally include both structure-based drug design (SBDD) and ligand-based drug design (LBDD) (Figure 2).29 With the rapid development of modern computer technology, structure-based virtual screening has become the most efficient and highly accurate method for the discovery of novel drug molecules and for conducting qualitative and quantitative studies on protein-ligand interactions. Since 2012, SBDD continues to aid drug discovery for GPCRs including adenosine, dopamine, and muscarinic receptors by identifying novel chemical scaffolds that target specific binding sites.30–32
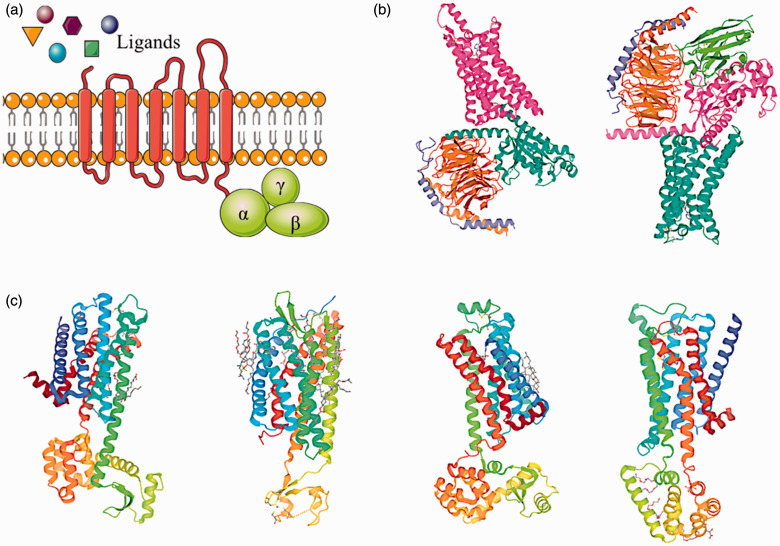
Figure 1.
Schematic diagram of a classic GPCR structure and representative three-dimensional GPCR-agonist, GPCR-antagonist complexes discussed in this review. (a) Schematic diagram of a GPCR seven-transmembrane structure. (b) Structures of the human adenosine A1 receptor-Gi2 protein complex bound to its endogenous agonist (PDB ID: 6D9H) and the adenosine A2A receptor bound to a mini-G heterotrimer (PDB ID: 6GDG). (c) Structures of the dopamine D2 receptor (PDB ID: 6CM4), P2Y1 receptor (PDB ID: 4XNV), β2 adrenergic receptor (PDB ID: 3NYA), and muscarinic M3 receptor (PDB ID: 4U15). (A color version of this figure is available in the online journal.)
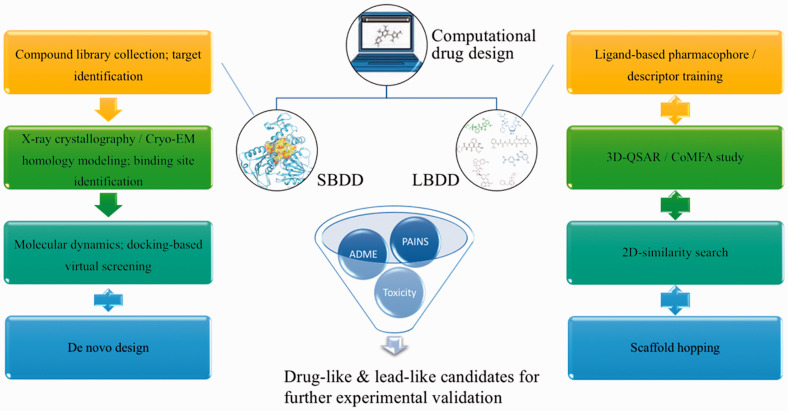
Figure 2.
General workflow of computational approaches, including SBDD and LBDD, commonly used for GPCR drug discovery. General procedures for SBDD start with a compound library collection and target identification. The structure of a target protein can be obtained by X-ray or cryo-EM and by homology modeling; subsequently, docking-based virtual screening, coupled with MD simulations, is performed to screen for hit compounds. Frequently used LBDD approaches include ligand-based pharmacophore models, 3D-QSAR, and CoMFA in combination with 2D-similarity searches. ML has been more commonly used in LBDD workflows to generate descriptors for diverse training models. Scaffold hopping serves as another complementary approach for generating new chemistry. (A color version of this figure is available in the online journal.)
Quantum chemistry, molecular mechanics (MM), and molecular dynamics (MD) are the basic theoretical calculation methods used in CADD. Applications of classic MD and advanced hybrid quantum mechanics (QM)/MM approaches enable atom-level simulations of biomolecular systems.33 During MD simulations, a virtual microscopy system is established, in which the motion of particles is in accordance with the classical laws of Newton's mechanics and the interactions between particles satisfy the superposition principle. Therefore, MD simulation, in combination with SBDD, is able to reflect the dynamics and flexibility of target proteins in nature in order to offer precise insights into molecular interactions between GPCRs and ligands.34 An increasing number of studies have shown that ligand-induced flexibility of GPCRs plays a vital role in numerous activation and signaling processes. Therefore, protein flexibility has been taken into account and incorporated in relaxation procedure and molecular docking of SBDD methods in software such as GOLD, Glide, and induced fit docking (IFD) of Schrödinger.35,36
In silico SBDD is the most practical technique when the three-dimensional structure of a disease-implicated drug target is known, as discussed above. SBDD includes two major strategies: molecular docking approaches and de novo ligand design. Since the 1960s, molecular docking has become the most widely used method in SBDD.37 Docking can provide theoretical calculations for target-ligand binding conformation and scores of their binding affinity, making it of great significance for both the initial screening of hit compounds and the computational analysis of lead compound binding patterns. Docking approaches comprise rigid docking from the classical drug-ligand “lock-and-key model”, and flexible docking from the later development of an “induced-fit model” and “conformational selection model”.38–40 Scoring functions for molecular docking mainly include force field-based, experience-based, knowledge-based, and descriptor-based scoring. The most commonly used D-score and G-score are based on Tripos force fields, while the Dock and Autodock algorithms are based on Amber force fields. Experience-based scoring functions include Chemscore and Glidescore.41 In recent years, machine learning (ML) has also been introduced into the scoring function by transforming protein-ligand interactions into descriptors that are further analyzed by random forest (RF), neural network, support vector machine (SVM), or Bayesian classification to establish effective scoring function models, such as NNScore and RF-Score.42
LBDD methods are utilized as alternatives to SBDD methods by exploiting the knowledge of known active and inactive molecules rather than the knowledge of structural information of a target protein. Therefore, LBDD methods are especially useful when the protein structure cannot be determined experimentally or predicted by computational methods. Even with the abundance of GPCR crystal and cryo-EM structures now available, LBDD remains in demand.43 The basic principle underlying LBDD is the assumption that similar molecules trigger similar effects, including biological actions and interaction with target proteins.44 A common practice in drug research is to explore similar molecules as a way to conveniently modulate certain characteristics, given a set of molecules that are effective towards a pharmaceutically relevant target.45 The most frequently used LBDD methods include ligand-based pharmacophores, molecular descriptors, and quantitative structure–activity relationships (QSAR).
Artificial intelligence (AI) and ML have also seen significantly extensive use on applications in quite a few research fields. Increased computational resources now allow researchers to develop more comprehensive databases and more effective algorithms targeting specific research applications. ML methods employ two basic learning strategies: supervised learning and unsupervised learning. These strategies accomplish diverse tasks, such as classification, regression, clustering, and dimension reduction.46 ML tasks for drug discovery pipelines require proper feature extraction methods and applicable algorithms. Major methods for feature extraction, in this regard, conversion of molecular structures into ML representation of chemical information, involve the graph-based methods, substructure mining, molecular descriptors, and molecular fingerprints.47 Algorithms are derived from SVM, convolutional neural networks, recurrent neural networks, and multilayer perceptrons (MLPs) as most frequently used in drug discovery workflows. Revolution in both SBDD and LBDD has been accomplished with the application of ML and additional novel strategies, and representative cases in recent years would be discussed below (Table 1).
Table 1.
Summary of representative cases discussed in this review regarding the recent application of CADD methodologies in GPCR structural studies and bioligand discovery.
| GPCR | Molecule/dataset | Research categories | Methodologies | Significance | Year |
|---|---|---|---|---|---|
| Dopamine D2 receptor; µ-opioid receptor | – | Molecular mechanism | All-atom MD | Metal ions detections at orthosteric site of ligand binding pocket | 2020 |
| P2Y1R | BPTU | Molecular mechanism; SBDD | Conventional MD; umbrella sampling; well-tempered and funnel-metadynamics | Investigations on membrane-embedded sites; identification of ligands to lipids-exposed sites | 2015/2018 |
| Dopamine receptor subtypes | Dopamine; 7-hydroxy-N,N-dipropyl-2-aminotetralin, etc. | Molecular mechanism | MD; homology modeling; molecular docking | New structural findings regarding binding modes and selectivity for DR subtypes | 2019 |
| Adenosine receptors; P2Y receptors | XAC; DPCPX; KW3902; ZMA; Z48; Z80, etc. | Molecular mechanism | Classic MD; metadynamics; temperature-accelerated MD; umbrella sampling | Assessment of complex stability and identification of alternative binding modes through induced fit | 2020 |
| Adenosine receptors A1AR; A2AAR | – | Molecular mechanism | Robust Gaussian accelerated molecular dynamics | Elucidation for detailed mechanism of specific AR-G protein coupling | 2019 |
| 5-HT2A; D2 subtypes | – | Molecular mechanism | ML-represented MD | Efficient computational analysis of “MD Big Data” and classification of ligands | 2019 |
| Olfactory receptor Olfr73 | ZINC database of 1.58 million candidates | SBDD | MD; molecular docking; PH4 search; interaction fingerprint | Accurate models with protein dynamics for OR agonist discovery | 2019 |
| Dopamine D2 receptor | ChEMBL10 | Molecular mechanism; SBDD | MD; homology modeling; molecular docking | Biased ligands-based design on specific amino acid–ligand contacts | 2018 |
| β-adrenergic receptor; muscarinic acetylcholine receptor | ZINC database | De novo; designfragment-based design | CIP-based fragment search; molecular docking; MD; binding free energy analysis | De novo design of GPCR ligands based on the relevant 3D structural information | 2019 |
| hAA2A; r5HT1A | 141,990 molecules in ChEMBL | Fragment-based design | Similarity-driven fragment-based approach | Evolutionary drug discovery approach for producing candidates | 2013 |
| Dopamine D4 receptor; sigma-1 receptor | 279,866 molecules in ChEMBL | LBDD | Adaptive fragment prioritization | Predictive quantitative poly-pharmacology models | 2014 |
| A2AAR; β1AR; β2AR; D3R; OX2 ; CXCR4, etc. | ZM241385; Suvorexant; IT1t, etc. | Molecular interactions | Fragment molecular orbital calculation | Balance between computational accuracy and speed; receptor-ligand binding interactions | 2016 |
| Adenosine A2A receptor | ChEMBL | De novo design | Reinforcement-learning with recurrent neural network | High diversity ligands generation with chemical and biological properties | 2019 |
| Odorant receptor OR51E1, etc. | Library of 258 odorants | LBDD | SVM-based virtual screening; QSAR; homology modeling | New perspective on the construction of QSAR models | 2018 |
| 5-HT2BR; 5-HT1BR | ChEMBL; MCule of 4.8 M molecules | LBDD; SBDD | NSFP-based ML model; molecular docking | Novel subtype-selective ligands identification | 2018 |
Molecular mechanisms of GPCR structure and signaling
GPCRs usually consist of three portions: an extracellular N-terminus, seven transmembrane α-helices with intracellular and extracellular loops, and an intracellular C-terminus. For this reason, they are also called hepta-helical receptors.9 The flexibility of GPCRs plays a vital role in various biological processes, including ligand recognition, orthosteric or allosteric modulation, activation, and signaling. GPCRs, when activated, control cellular signal transduction across membranes.48 The signal is initiated from the extracellular side with incorporation of endogenous ligand molecules that stabilize the active conformation of the receptor, leading to cellular response through dimerization with intracellular G proteins or other intracellular proteins.49 The activation-related conformational changes of GPCRs have been clarified by recent studies, and this has widen our understanding of structural basis of the interactions between GPCRs and their partners.50 Upon activation, C-terminal α-helix of G protein α-subunit interacts with the cavity on cytoplasmic side of GPCRs with outward movement of TM6 region.
Rapid progress in GPCR structural biology in recent years has led to an unprecedented elucidation of detailed structural basis of GPCR activation process. Numerous structures of GPCR-G protein or GPCR-arrestin complexes have now been deciphered by X-ray crystallography and cryo-EM in recent years. More than 60 unique GPCR structures and 13 structures of GPCRs bound to a nucleotide free G protein heterotrimer, covering 3 different Gα families, have been determined to date.51 Nonetheless, a variety of challenges remain in achieving high-resolution active or active-intermediate states of GPCR activation and ligand association process.52 Besides, membrane is usually mimicked by use of detergents in vitro which limits precise representation of physiological environment. To address mechanistic questions at an atomic level, the integration of structural biology with computational methodologies is essential for exploring the structural dynamics and molecular mechanisms of GPCRs with multiple conformational states.53 MD simulation for ligand-target complexes takes protein flexibility into consideration and can account for explicit solvent, ions, and membranes in the biosystem. Previously, researchers have summarized how atomic dynamics modeling revealed transient states with high energy barrier of ligand binding process, provided insights into effects of receptor mutations on ligand affinity and kinetics and helped understand target druggability.54–56 Therefore, MD simulations have been increasingly applied to GPCRs to elucidate the conformational changes of key residues and motifs, as well as to compute diverse binding modes, target specificity, and binding kinetics.57
Chan et al. probed the existence of sodium ions in the dopamine D2 receptor (D2R) using all-atom MD simulations in 2020.58 As we know, GPCRs are usually regulated by “allosteric site” metal ions which render the GPCR inactive. In the case of D2R, an allosteric sodium ion locates next to D802.50 in apo D2R and decreases the affinities of agonists, thereby leading to coupling of the receptor to the Gi/o protein. This in turn, inhibits adenylyl cyclase, while enhancing the affinities for some antagonists. Chan’s use of computational and pharmaceutical methods showed for the first time that a sodium ion could be located at the “orthosteric site” of the ligand-binding pocket and enable coordination of a polar residue with a specific agonist molecule. First, a complex of D2R and a potent agonist MLS1547 was computationally constructed by docking. Interestingly, MD simulations revealed an additional sodium ion in the orthosteric ligand binding site of the extracellular region, forming coordination with D2R and the agonist molecule at 1.0 − 3.2 μs time scale. These results were also observed for μ-opioid receptor and further verified by high-resolution crystal structure determination and biochemical experiments. In addition, by enhancing the interaction at the particular binding site, researchers were able to modify candidate molecules and increase their activities by 16-fold. This strategy, together with the workflow, has opened up new opportunities at a mechanism level of GPCR signaling characteristics and optimization of GPCR ligand molecules.
Understanding the incorporation of GPCR ligands to membrane-embedded sites remained a challenge, as substitution of detergents for membrane in crystallization procedures limits the precise representation of physiological membrane environment. BPTU is an antagonist for purinergic P2Y1 receptor (P2Y1R). It targets a special extra-helical site that locates in-between the membrane and the protein.26 In 2018, Yuan et al. conducted all-atom simulations with models of BPTU and a POPC bilayer. The simulation workflow involved both conventional MD and multiple enhanced sampling methods, including umbrella sampling, well-tempered metadynamics as well as funnel-metadynamics simulations.59 Researchers explored molecular mechanism underlying the allosteric binding of BPTU to the extra-helical site of P2Y1R, and found that BPTU prefers to partition into the interface of lipophilic region, before interacting with the second extracellular loop of P2Y1R. The study further provided accurate binding free energy calculation which was in remarkable agreement with experimental data. Yuan’s work offered a reference for investigations on other membrane-embedded sites, and identification of ligands to lipid-exposed sites of membrane proteins.
Homology modeling, fold recognition, de novo folding, and MD simulations are major approaches for building conformational models of GPCRs. Bueschbell et al., in 2019, applied homology modeling and MD simulation to construct robust conformational models of all the dopamine receptor (DR) subtypes and further performed docking for structurally diverse ligands. These experiments revealed new structural findings regarding the binding modes and selectivity for DR subtypes.60 Salmaso et al., in a review article, provided a detailed discussion of recent MD studies of purinergic GPCRs, including well-known adenosine receptors and P2Y receptors.61 This article indicated that enhancement of traditional MD simulations algorithms has enabled researchers to explore more complicated and long-timescale phenomena, including receptor activation, recognition, and dissociation pathways in GPCR biosystems at an atomistic level. Furthermore, Wang et al. employed all-atom simulations by a robust Gaussian accelerated molecular dynamics (GaMD) method on adenosine receptors (ARs), including subtypes of A1AR and A2AAR, and deciphered detailed mechanism of specific AR-G protein coupling.62 GaMD simulations and free energy calculation revealed preferential coupling of A1AR-Gi and A2AAR-Gs complexes, which were highly consistent with experimental findings.
ML algorithms have also been integrated with traditional MD simulation, as a great mass of data accumulates gradually from large-scale simulations. Meanwhile, automated de novo design using deep learning (DL) methods has been accomplished via numerous automated compound generators and selection operators in recent years.63 For instance, Plante et al., in 2019, described a novel approach that was based on transforming the analysis of GPCR function-related, ligand-specific differences encoded in the MD simulation trajectories into a representation recognizable by state-of-the-art DL object recognition technology.64 This method was subsequently used on the serotonin 5-HT2A receptor and D2 subtypes for high-accuracy identification of the pharmacological classification of ligands. These results presented a feasible framework for efficient computational analysis of “MD Big Data” collected to understand ligand-specific GPCR activities.
Oligomer-specific drug design seems to be an increasing need, as evidence accumulates that GPCRs tune their functions through oligomer formation and protein–protein interactions. While structural information about GPCR oligomers becomes more demanded, technical obstacles to crystallization and biochemistry are also amplified. In 2014, Schonenbach et al. provided an overview of mechanistic and functional models for GPCR oligomers, and perspectives on emerging techniques to characterize GPCR oligomers.65 In this case, MD simulations become a powerful tool to predict oligomer interfaces and analyze their stabilities, and have been applied to multiple GPCRs including rhodopsin, β2AR, β1AR, and μ-opioid receptors. Overall, studies in the past few years have provided more insights into practicability of MD simulations in various biological processes regarding GPCRs, filling the gap between protein structures and functions.66 Improvements in MD algorithms enable the observation of finely tuned conformational changes that are critical in intracellular signal transductions.67 Application of these enhanced approaches would significantly transform the efforts on current drug discovery for GPCR monomers and oligomers.
Receptor-based rational design
Structure-based rational design has played an outstanding role over the past decades in the discovery of bioactive ligands for GPCRs and has identified a number of small molecules with therapeutic significance. Structural information is essential for SBDD of novel bioactive molecules. SBDD has benefited from the rapid development and technology breakthroughs in X-ray crystallography and cryo-EM; therefore, the search for potent GPCR agonists or antagonists as promising lead compounds has now entered a new phase. For example, using GPCR stabilization and SBDD technologies, small molecule AZD4635 was derived as an antagonist of the immune checkpoint target A2AR. It has entered a Phase II clinical trial in 2019 for the treatment of advanced solid tumors.68 While rapidly emerging information for GPCR has enhanced the effectiveness and accuracy of rational ligand design process, multiple computational tools become available and can be utilized simultaneously. Ligand docking, free energy calculation, and de novo ligand design are most practicable approaches for SBDD. Free energy calculation methods, for instance, thermodynamic integration and free energy perturbation, have provided additional support for rigorous prediction of GPCR-ligand binding affinity.69,70 Meanwhile, with the frequent application of molecular docking for drug discovery, an increasing number of easy-to-use bioinformatics tools for homology modeling and docking have become available online in recent years as complements to the classic software and packages, such as Autodock, GLIDE, and Surflex-Dock (Table 2).71–77
Table 2.
Representative computational and bioinformatics tools for structure-based virtual screening published in recent years.
| Platform | Functions and features | Link | Year |
|---|---|---|---|
| EasyVS | Molecule library construction and docking | http://biosig.unimelb. edu.au/easyvs/ | 2020 |
| HawkDock | Prediction and analysis of protein–protein complex based on docking and MM/GBSA | http://cadd.zju.edu.cn/hawkdock/ | 2019 |
| DockThor 2.0 | Protein-ligand docking and binding mode prediction utilizing high-performance platform and supercomputer | http://www.dockthor.lncc.br/ | 2017 |
| pepATTRACT | Large-scale protein–peptide docking | http://bioserv.rpbs.univ- paris-diderot.fr/services/pepATTRACT/ | 2017 |
| AMMOS2 | Protein–ligand–water complexes refinement via molecular mechanics | http://drugmod.rpbs.univ-paris-diderot.fr/ammosHome.php/ | 2017 |
| PPI3D | Template-based modeling and search for homologous protein complexes | http://bioinformatics.lt/software/ppi3d/ | 2017 |
| SEABED | Receptor preparation, library editing, flexible ensemble docking, hybrid docking and QSAR | http://www.bsc.es/SEABED/ | 2015 |
| MTiOpenScreen | Small molecule docking for user-defined binding site or blind docking | http://bioserv.rpbs.univ-paris-diderot.fr/services/MTiOpenScreen/ | 2015 |
Olfactory receptors (ORs) are one of the major members in the GPCR superfamily. An increasing number of high-resolution 3D structures of non-olfactory receptors has accelerated the rational drug design and the understanding of receptor-ligand interactions in recent year. By contrast, little is known about ORs: the cognate agonists of most ORs have not yet been identified and discovery of their agonists remains challenging.78,79 For agonist discovery, protein dynamics are critical for the development of accurate models. Therefore, adoption of MD simulation in SBDD projects is encouraged for OR ligand discovery. Several computational studies on ligand prediction of ORs have been published in recent years (Table 3). In 2019, Yuan et al. provided a proof-of-principle study for identifying novel therapeutic OR agonists using virtual screening with MD simulations.80 They constructed a three-dimensional structure model of olfactory receptor Olf73 by homology modeling and optimized structural conformation of the initial model by MD simulation. A smaller but more flexible binding pocket of Olfr73 was revealed common to most of the known OR agonists, rather than the binding pockets of typical non-ORs. Moreover, virtual screening of a library of 1.58 million compounds against Olfr73 was conducted; 25 predicted Olfr73 agonists were tested by cell-based assays, of which 17 compounds were validated as effective with a hit rate of 68%. Further interaction fingerprint analysis for these newly found agonists indicated much fewer polar interactions between the OR and ligands than was observed with other GPCRs, as well as a limitation of this pocket size for agonist binding.
Table 3.
Computational studies for ligand prediction of olfactory receptors in recent years.
| Olfactory receptors | Representative known ligands | Molecule/dataset for computational studies | Research categories and methodologies | Year |
|---|---|---|---|---|
| OR51E1 | Eugenyl acetate; 2,4-dinitrotoluene; methyl furfuryl disulfide | Library of 128 compounds from literature | LBDD. SVM-based virtual screening; QSAR; homology modeling | 2018 |
| OR1A1 | Ethyl phenylacetate; trans-anethole | Library of 315 compounds from literature | LBDD. SVM-based virtual screening; QSAR; homology modeling | 2018 |
| OR2W1 | Octanal; benzophenone; eugenyl acetate | Library of 274 compounds from literature | LBDD. SVM-based virtual screening; QSAR; homology modeling | 2018 |
| MOR256-3 | Benzaldehyde; 2,3-butanediol; coumarin | Library of 73 compounds from literature | LBDD. SVM-based virtual screening; QSAR; homology modeling | 2018 |
| OR1G1 | Nonanal; 9-decen-1-ol; camphor | Library of 173 compounds from literature | LBDD. SVM; RF; naı ¨ve Bayes; neural network | 2018 |
| Olfr73 | Isoeugenol; p-isobutylphenol | ZINC database of 1.58 million molecules | SBDD. MD; molecular docking; PH4 search; interaction fingerprint analysis | 2019 |
| MOR42-3 | α-hexyl cinnamaldehyde | Library of 574 odorants | SBDD. Homology modeling; molecular docking; free energy calculation | 2014 |
Characterization of biased signaling and identification of biased agonists for GPCR now remain challenging, even with the abundance of available crystal structures. Biased agonists represent ligands that induce distinct active conformations of a receptor, thereby activating specific subsets of its functional signaling profiles.81 Understanding of the mechanisms underlying biased agonism of GPCRs and characterization of ligands has therefore been increasing in recent years. Biased agonists targeting GPCRs, such as the angiotensin type I receptor and μ-opioid receptor, have reached the late stages of clinical development, providing potential therapeutic benefits including higher efficiencies and reduced adverse effects.82,83 Challenges in studying biased signaling underlie the limitations in understanding the complexity of GPCR functionality and in detecting specific types of signaling dynamics.84 Several recent studies have confirmed that SBDD represents a powerful strategy for identifying novel scaffolds of biased agonists (Table 4).85–90 In 2017, Man¨nel et al. performed a SBDD study on functionally selective D2R ligands in order to pursue fine-tuning of functional receptor activities.87 The structure of D2R was predicted by homology modeling using the crystal coordinates of the D3R subtype and molecular docking of known functionally selective ligands indicated the ligand-receptor interactions within the orthosteric site and an extension into a secondary pocket. A virtual library with about 13,000 compounds was screened based on that model, and 16 partial agonists were discovered out of the 18 top-ranked compounds. Mccorvy et al., in 2018, presented an approach that applied a combination of homology modeling, docking, and MD simulation to translate GPCR structural data into β-arrestin-biased ligands for aminergic GPCRs.88 In that work, the researchers used D2R as a model system to identify GPCR–ligand contacts that mediate biased signaling, and they identified specific amino acid–ligand contacts at transmembrane helix 5 and extracellular loop 2 that are responsible for Gi/o and β-arrestin signaling. They further used specific templates targeting those residues to develop biased ligands. This work illustrated a successful combined strategy for designing biased ligands, based on a combination of computational and biochemical approaches, and provided a good example of leveraging GPCR structures to create biased drugs. With increasing clinical development of biased agonists ongoing for a variety of indications, we believe that future drug design will continue to assess ligand bias in order to develop safer and perhaps more effective medications.
Table 4.
Successful cases for the discovery of biased agonists for GPCRs in recent years.
| GPCR | Representative biased agonist | Molecule/dataset for computational studies | Research categories and methodologies | Year |
|---|---|---|---|---|
| MT1/MT2 | 4-phenyl-2-propionamidotetralin | Library of 8.4 million fragment-like and lead-like compounds | SBDD. Molecular docking | 2020 |
| D2R | UNC0006; aripiprazole | Library of about 13000 compounds | SBDD. Homology modeling; molecular docking | 2017/2018 |
| 5-HT2AR | 2,5-dimethoxy-4- nitrophenethylamine | 3-(aminoethyl)1-methylindol-5-ol; 5-methyltryptamine; 5-Nitro-1H-indole-3-ethanamine | SBDD. MD; molecular docking; ligand-residue interaction analysis | 2015 |
| β2AR | BI-167107 | ZINC library of 3.4 million lead-like and fragment-like molecules | SBDD. Homology modeling; molecular docking | 2013 |
De novo small molecule design is another frequently used SBDD strategy. The basic principle of de novo design is to create novel chemical entities with specific properties from scratch or to search the same space for new structures with drug properties. By using structural information of either target information or structure–activity relationship data, de novo design of a drug offers an efficient and intellectually appealing alternative to molecular docking of a large compound database through the use of either the structural information about the target or the structure–activity relationship data.91 This provides a broader exploration of chemical space to identify novel scaffolds in a cost- and time-efficient manner. Today, with the growing capabilities in chemical synthesis and computational speed, de novo design has become increasingly in demand by researchers to deliver attractive ideas for chemical generation and drug discovery.92 Numerous algorithms have been developed to improve the performance of de novo design, especially with the re-emergence of ML in recent years. In 2019, Li et al. presented de novo design for GPCR ligands based on relevant 3D structural information.93 They performed a fragment-based workflow by first extracting the characteristic interaction patterns (CIPs) on the binding interfaces between the GPCRs and ligands. They further employed these CIPs to search GPCR ligands for chemical fragments, which would form similar interaction patterns with GPCRs. The selected chemical fragments were further assembled into complete molecules using the AutoT&T2 software. This strategy was well validated in the cases of the β-adrenergic receptor and muscarinic acetylcholine receptor and identified a total of 15 and 22 compounds, respectively, as active antagonists for these two receptors. Further MD simulations and binding free energy analyses were performed to explore the key interactions between those active compounds and their targets. The described workflow presented an effective fragment-based design method for the β-adrenergic receptor and the muscarinic acetylcholine receptor based on CIP analysis. With the increasing improvements in de novo design approaches, we should be aware of the remaining challenges, such as precise description of molecular fingerprints or scoring functions. The combination of ML-driven generative molecular design models with novel algorithms for identification of activity-specific fragments could represent promising directions for future molecular discovery and optimization.
Ligand-based rational design
Typically, LBDD serves as a valuable alternative for agonist and antagonist discovery for GPCRs, especially for GPCRs that currently lack three-dimensional structural information. LBDD employs approved drugs or known active molecules of interest as references and sets up pharmacophore models to discover novel chemical structures.94 Several GPCR-targeting compounds that are derived by LBDD workflow are currently under clinical trials. For instance, SEP-363856 is an antipsychotic targeting 5-HT1A receptor.95 To achieve better binding affinity, it was optimized based on QSAR model with a Ki value of less than 1 μM. The agent has entered Phase III clinical study in 2019 for the treatment of adults and adolescents with schizophrenia. These days, with the re-emergence of AI and the accumulated databases, ML approaches have contributed greatly to GPCR cheminformatics for the improvement of chemical descriptor calculations and classification algorithms. This definitely increases our confidence in the integration of LBDD and SBDD workflows that combine each of their strengths as discussed below.
GPCR SARfari database is an integrated chemo-genomics workbench for GPCR studies and drug discovery. In 2013, Kawai et al. proposed a similarity-driven fragment-based evolutionary approach for producing candidate molecules for drug discovery.96 In that study, bioactive molecules in the GPCR SARfari database were used to prepare a fragment library. Ligand design for the hAA2A and r5HT1A receptors was carried out to verify the feasibility of this approach. In 2014, Reutlinger et al. presented a de novo design method that used adaptive fragment prioritization.97 They developed a predictive quantitative poly-pharmacology model for 640 human drug targets based on publicly available structure–activity data. Using this model, they obtained novel subtype-selective and multitarget-modulating dopamine D4 antagonists, as well as ligands selective for the sigma-1 receptor, thereby proving the applicability of using adaptive building blocks and fragment prioritization. Likewise, the classic fragment molecular orbital (FMO) method proposes an excellent solution that balances accuracy and speed. It offers a considerable speed-up, since quantum mechanics approaches are often too computationally expensive, and it also has the potential to explore key interactions and selectivity that would otherwise be hard to detect.98,99 For example, Bodkin et al. described how FMO has been applied to the analysis of 18 GPCR-ligand crystal structures representing different branches of the GPCR genome.100,101 This approach could provide more comprehensive receptor-ligand binding interactions, including those that are often omitted from structure-based descriptions like hydrophobic interactions, or nonclassical hydrogen bonds, and would shed light on both LBDD and SBDD for these receptors.
Meanwhile, ML algorithms have been gradually introduced into the LBDD workflow. For instance, application of DL provides a new perspective on the construction of QSAR models or the generation of novel chemical structures.102,103 This requires the conversion of molecular structures into chemical information that can be processed computationally. Several studies have been presented on the automatic extraction of descriptors from chemical structures using neural network models such as extended connectivity fingerprint (ECFP) and Mol2Vec, as well as autoencoder models like DruGAN approaches.104–106 DrugEx is a recurrent neural network generator trained through reinforcement learning for de novo drug design and has been applied to ligand discovery against the adenosine A2A receptor.107 In another study, Rataj et al. employed a hierarchical combination of ligand-based ML classification and structure-based molecular docking methods to discover novel compounds with 5-HT2BR versus 5-HT1BR selectivity.108 A neighboring substructure fingerprint (NSFP)-based ML model was built using in vitro activity data for human 5-HT1BR and 5-HT2BR receptors obtained from ChEMBL. The activity and selectivity classifiers for 2B were developed and the final models were selected based on the highest acquired Matthews correlation coefficient values. This ML-based classification was subsequently combined with complementary docking workflows and applied to a MCule database of 4.8 M molecules. Three hits were identified with nanomolar affinity and over 10-fold selectivity.
Application of ML algorithms in LBDD also addresses the longstanding challenge of predicting the activity of chemicals for odorant receptors. The olfactory receptor database (ORDB) offers an integration of genomic and proteomic information related to ORs, as well as detailed ligand molecules that have been experimentally shown to interact with and activate ORs.109 This provides a good basis for further ligand-based computational studies on ORs. In 2018, Bushdid et al. performed a study to predict the activity of chemicals for a given odorant receptor using a ML algorithm.110 The activities of 258 chemicals on odorant receptor OR51E1 were virtually screened using 4884 chemical descriptors as inputs and two novel agonists were identified and validated by in vitro experiments. The SVM-based protocol was further assessed on other odorant receptors including OR1A1, OR2W1, and MOR256-3, and the resulting hit rates for novel agonists were around 39–50%. This was one of the first successful cases of applying ML algorithms to agonist discovery of ORs. Inspired by the SVM-based prediction, another case study used diverse ML methods to identify potential agonists of olfactory receptor OR1G1 47. Three classical ML algorithms, including SVM, RF, and naı ¨ve Bayes, as well as a neural network-based method, were employed. After selecting the best prediction results, the top ranked compounds were characterized as pyrazines, benzene-containing ketones, and esters.
Another important application to address is the prediction of drug-induced cardiotoxic effects. Blockade of the human Ether-a-go-go Related-Gene (hERG) potassium channel has historically been a barrier to drug development, as reduced functionality of the hERG channel causes QT prolongation, which may lead to severe cardiotoxicities, such as cardiac arrhythmia.111 Notorious cases of several approved drugs, including astemizole, terfenadine, and cisapride that have been withdrawn from the market due to their cardiotoxic effects, addressed the importance of evaluating the hERG-blocking activity of drug candidates at the hit selection stage.112,113 Therefore, computational approaches have been developed to predict potential hERG blockage of preclinical drug candidates as a way to reduce the risk of drug attrition. Diverse structure-based and ligand-based approaches have shed more light on the molecular basis of drug-channel interactions.114–118 Here, we mainly discuss two ligand-based examples. Chemi et al. generated a ligand-based pharmacophore workflow, followed by development and validation of a 3 D-QSAR model, in pursuit of a fast and reliable in-house computational tool for estimating hERG activity.119 A total of 730 molecules were used for the study and five features (two aromatic rings, one hydrogen-bond acceptor, one hydrophobic site, and one positive ionizable function) comprised the pharmacophore model. The sequential 3D-QSAR model developed with a set of 421 compounds proved to be predictive with the ROC of 0.96. The performances in terms of sensitivity and negative predictive value have been improved using ML-based approaches in recent years.120,121 For instance, Ryu et al. in 2020, proposed a computational framework named DeepHIT, which contains different DL models that produce fewer false negative predictions. A dataset of 6632 hERG blockers and 7808 non-blockers was generated and three independent DL models were trained.112 DeepHIT presented a higher accuracy, MCC, sensitivity, and NPV than previous prediction tools. As a proof-of-concept study, these researchers also identified novel urotensin II receptor antagonists without hERG-blocking activity derived from a previously reported UT antagonist with a strong hERG-blocking activity. In summary, these computational tools like DeepHIT have contributed greatly to rational design and optimization in the early stages of drug discovery and development.
Conclusions
GPCR proteins have extensive physiological roles; therefore, they are a prominent target category for pharmaceuticals. They have been pursued as major therapeutic targets for decades by virtue of their contributions in cellular communications. Deeper understanding of GPCR targets and their corresponding ligands today has resulted in increasing efforts to integrate all these new information into computer-derived models to further benefit the drug discovery process. In this review, we provide an update on state-of-the-art computational approaches for GPCR drug discovery from the aspect of MD simulation, structure-based and ligand-based rational drug design. A wide variety of novel algorithms or workflows has been developed and applied to the GPCR ligand discovery process, thereby guiding further drug design for analogous targets. In addition, ML methods are now extensively used to build more accurate computational models as data on drug discovery accumulate.
Enhancement of computing power provided by new technologies has made MD simulation a helpful tool in molecular modeling and SBDD. Though numerous GPCR conformations and complexes have been deciphered by X-ray crystallography and cryo-EM in recent years, difficulties still remain in obtaining active states, in observing finely tuned conformational changes, and in decoding molecular mechanisms such as allostery for specific GPCRs. MD simulations, together with enhanced sampling techniques, have continuously aided our understanding of the detailed and dynamic processes how GPCR structures transduce physiological signals into diverse cellular responses. But there are remaining challenges in accurate simulation for the controlling systems of GPCR activation, as well as the prediction of thermodynamic and kinetic properties during ligand binding to GPCRs. Improved exercises in GPCR dynamics could further contribute to the discovery and development of more selective and effective drug molecules, including agonists.
The use of SBDD and LBDD methodologies has dramatically increased in the last decades and contributed greatly to GPCR rational drug design. SBDD has benefited from the rapid accumulation of three-dimensional structures, while LBDD develops with the evolution of database technology. Both of these protocols facilitate the discovery of novel ligand molecules. The integration of MD simulations also offers an outlet for incorporating protein flexibility into the SBDD workflow of GPCR-targeted ligands. Moreover, free energy calculation has offered significant potential in accurate evaluation of absolute and relative binding affinities. Programs that could balance between speed and accuracy would be promising as a general procedure for GPCR-targeting SBDD studies. Nonetheless, the rational design of GPCR agonist ligands remains arduous and requires extensive effort on elucidating transient states and conformational changes of ligand binding. Emergence of novel AI-based methodologies has opened up new research avenues for drug discovery of GPCRs, especially for LBDD where molecular feature presentation proves to be the key step. With the boom of molecular databases and ML algorithms, LBDD methods would undoubtedly take the identification of small molecule GPCR ligands to the next step. However, an important point to note is that, even as computational methods for drug design are becoming more and more advanced, human assessment based on expert experience of computer-generated outcomes is still essential on a case-by-case basis. Another challenge posed here is the interpretation of AI-generated results. It is usually challenging for chemists and biologists to understand the direct output of what a deep neural network has learned after the training process. As data further accumulate, this gap may continue to grow and could impede further use of AI models in drug discovery and development. Besides, it should be noted that no documented examples of licensed drugs derived from SBDD are currently available; however, preclinical and clinical evidence, such as the development of AZD4635, has been accumulating over time that would support an impact of structural biology on drug design and development. Overall, the coupling of SBDD and LBDD could be particularly appealing in the future, especially when coupled with appropriate employment of known databases and annotations.
Wide distribution and significant roles of GPCRs in cellular physiology create a continuously increasing demand today for novel GPCR modulators. This review highlights the recent progress and updates on computational methods for the discovery of GPCR bioactive ligands. The future is expected for GPCR drug discovery through rapid development of parallel techniques that include both experimental methodologies and computational strategies. However, no magic shortcut exists for GPCR drug discovery. Scientists often state that computational drug design is more of an art than science. The diversity of GPCR structures and conformations constantly poses new challenges for drug discovery, and no procedural approaches yet exists that generally apply to all GPCRs. Computational discovery of each novel drug candidate requires an in-depth understanding of the complexity of a specific receptor-ligand pair, while requiring rational thinking with a receptive frame of mind to promote a multidisciplinary partnership. With the continuing effort and enhanced strategies, it is anticipated that increasing success cases for GPCR drug discovery driven by more advanced computational techniques, will come to pass in the forthcoming years.
Footnotes
AUTHORS’ CONTRIBUTIONS: SZ and MW wrote the manuscript. ZH and JA supervised the work and revised the manuscript.
DECLARATION OF CONFLICTING INTERESTS: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
FUNDING: This work was supported in part by grant from the National Institutes of Health (GM57761).
ORCID iD: Siyu Zhu https://orcid.org/0000-0001-5352-2273
References
- 1.Munk C, Mutt E, Isberg V, Nikolajsen LF, Bibbe JM, Flock T, Hanson MA, Stevens RC, Deupi X, Gloriam DE. An online resource for GPCR structure determination and analysis. Nat Methods 2019; 16:151–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pierce KL, Premont RT, Lefkowitz RJ. Seven-transmembrane receptors. Nat Rev Mol Cell Biol 2002; 3:639–50 [DOI] [PubMed] [Google Scholar]
- 3.Congreve M, de Graaf C, Swain NA, Tate CG. Impact of GPCR structures on drug discovery. Cell 2020; 181:81–91 [DOI] [PubMed] [Google Scholar]
- 4.Southan C, Sharman JL, Benson HE, Faccenda E, Pawson AJ, Alexander SPH, Buneman OP, Davenport AP, McGrath JC, Peters JA, Spedding M, Catterall WA, Fabbro D, Davies JA. Nc-Iuphar. The IUPHAR/BPS guide to PHARMACOLOGY in 2016: towards curated quantitative interactions between 1300 protein targets and 6000 ligands. Nucleic Acids Res 2016; 44:D1054–D68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Roth BL, Kroeze WK. Integrated approaches for genome-wide interrogation of the druggable non-olfactory G protein-coupled receptor superfamily. J Biol Chem 2015; 290:19471–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sriram K, Insel PA. G protein-coupled receptors as targets for approved drugs: how many targets and how many drugs? Mol Pharmacol 2018; 93:251–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hauser AS, Attwood MM, Rask-Andersen M, Schioth HB, Gloriam DE. Trends in GPCR drug discovery: new agents, targets and indications. Nat Rev Drug Discov 2017; 16:829–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Santos R, Ursu O, Gaulton A, Bento AP, Donadi RS, Bologa CG, Karlsson A, Al-Lazikani B, Hersey A, Oprea TI, Overington JP. A comprehensive map of molecular drug targets. Nat Rev Drug Discov 2017; 16:19–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Doze VA, Perez DM. G-protein-coupled receptors in adult neurogenesis. Pharmacol Rev 2012; 64:645–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wu F, Song G, de Graaf C, Stevens RC. Structure and function of peptide-binding G protein-coupled receptors. J Mol Biol 2017; 429:2726–45 [DOI] [PubMed] [Google Scholar]
- 11.Cherezov V, Rosenbaum DM, Hanson MA, Rasmussen SG, Thian FS, Kobilka TS, Choi HJ, Kuhn P, Weis WI, Kobilka BK, Stevens RC. High-resolution crystal structure of an engineered human beta2-adrenergic G protein-coupled receptor. Science 2007; 318:1258–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Caffrey M, Li D, Dukkipati A. Membrane protein structure determination using crystallography and lipidic mesophases: recent advances and successes. Biochemistry 2012; 51:6266–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Munk C, Isberg V, Mordalski S, Harpsoe K, Rataj K, Hauser AS, Kolb P, Bojarski AJ, Vriend G, Gloriam DE. GPCRdb: the G protein-coupled receptor database – an introduction. Br J Pharmacol 2016; 173:2195–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kowalsman N, Niv MY. GPCR & company: databases and servers for GPCRs and interacting partners. Adv Exp Med Biol 2014; 796:185–204 [DOI] [PubMed] [Google Scholar]
- 15.Zhao Q, Wu BL. Ice breaking in GPCR structural biology. Acta Pharmacol Sin 2012; 33:324–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rouck JE, Krapf JE, Roy J, Huff HC, Das A. Recent advances in nanodisc technology for membrane protein studies (2012-2017). FEBS Lett 2017; 591:2057–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Draper-Joyce CJ, Khoshouei M, Thal DM, Liang YL, Nguyen ATN, Furness SGB, Venugopal H, Baltos JA, Plitzko JM, Danev R, Baumeister W, May LT, Wootten D, Sexton PM, Glukhova A, Christopoulos A. Structure of the adenosine-bound human adenosine A1 receptor-Gi complex. Nature 2018; 558:559–63 [DOI] [PubMed] [Google Scholar]
- 18.Maeda S, Qu Q, Robertson MJ, Skiniotis G, Kobilka BK. Structures of the M1 and M2 muscarinic acetylcholine receptor/G-protein complexes. Science 2019; 364:552–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Garcia-Nafria J, Nehme R, Edwards PC, Tate CG. Cryo-EM structure of the serotonin 5-HT1B receptor coupled to heterotrimeric go. Nature 2018; 558:620–3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gao Y, Hu H, Ramachandran S, Erickson JW, Cerione RA, Skiniotis G. Structures of the rhodopsin-transducin complex: insights into G-protein activation. Mol Cell 2019; 75:781–90.e3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Garcia-Nafria J, Lee Y, Bai X, Carpenter B, Tate CG. Cryo-EM structure of the adenosine A2A receptor coupled to an engineered heterotrimeric G protein. Elife 2018; 7:e35946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Koehl A, Hu H, Maeda S, Zhang Y, Qu Q, Paggi JM, Latorraca NR, Hilger D, Dawson R, Matile H, Schertler GFX, Granier S, Weis WI, Dror RO, Manglik A, Skiniotis G, Kobilka BK. Structure of the micro-opioid receptor-Gi protein complex. Nature 2018; 558:547–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hauser AS, Gloriam DE, Brauner-Osborne H, Foster SR. Novel approaches leading towards peptide GPCR de-orphanisation. Br J Pharmacol 2020; 177:961–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Granier S, Kobilka B. A new era of GPCR structural and chemical biology. Nat Chem Biol 2012; 8:670–3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thorsen TS, Matt R, Weis WI, Kobilka BK. Modified T4 lysozyme fusion proteins facilitate G protein-coupled receptor crystallogenesis. Structure 2014; 22:1657–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang D, Gao ZG, Zhang K, Kiselev E, Crane S, Wang J, Paoletta S, Yi C, Ma L, Zhang W, Han GW, Liu H, Cherezov V, Katritch V, Jiang H, Stevens RC, Jacobson KA, Zhao Q, Wu B. Two disparate ligand-binding sites in the human P2Y1 receptor. Nature 2015; 520:317–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wacker D, Fenalti G, Brown MA, Katritch V, Abagyan R, Cherezov V, Stevens RC. Conserved binding mode of human beta2 adrenergic receptor inverse agonists and antagonist revealed by X-ray crystallography. J Am Chem Soc 2010; 132:11443–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Leelananda SP, Lindert S. Computational methods in drug discovery. Beilstein J Org Chem 2016; 12:2694–718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sliwoski G, Kothiwale S, Meiler J, Lowe EW., Jr. Computational methods in drug discovery. Pharmacol Rev 2014; 66:334–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Congreve M, Andrews SP, Dore AS, Hollenstein K, Hurrell E, Langmead CJ, Mason JS, Ng IW, Tehan B, Zhukov A, Weir M, Marshall FH. Discovery of 1,2,4-triazine derivatives as adenosine A(2A) antagonists using structure based drug design. J Med Chem 2012; 55:1898–903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Christopher JA, Aves SJ, Bennett KA, Dore AS, Errey JC, Jazayeri A, Marshall FH, Okrasa K, Serrano-Vega MJ, Tehan BG, Wiggin GR, Congreve M. Fragment and structure-based drug discovery for a class C GPCR: discovery of the mGlu5 negative allosteric modulator HTL14242 (3-chloro-5-[6-(5-fluoropyridin-2-yl)pyrimidin-4-yl]benzonitrile). J Med Chem 2015; 58:6653–64 [DOI] [PubMed] [Google Scholar]
- 32.Wang S, Wacker D, Levit A, Che T, Betz RM, McCorvy JD, Venkatakrishnan AJ, Huang XP, Dror RO, Shoichet BK, Roth BL. D4 dopamine receptor high-resolution structures enable the discovery of selective agonists. Science 2017; 358:381–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hollingsworth SA, Dror RO. Molecular dynamics simulation for all. Neuron 2018; 99:1129–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee Y, Lazim R, Macalino SJY, Choi S. Importance of protein dynamics in the structure-based drug discovery of class a G protein-coupled receptors (GPCRs). Curr Opin Struct Biol 2019; 55:147–53 [DOI] [PubMed] [Google Scholar]
- 35.Verdonk ML, Cole JC, Hartshorn MJ, Murray CW, Taylor RD. Improved protein-ligand docking using GOLD. Proteins 2003; 52:609–23 [DOI] [PubMed] [Google Scholar]
- 36.Sherman W, Day T, Jacobson MP, Friesner RA, Farid R. Novel procedure for modeling ligand/receptor induced fit effects. J Med Chem 2006; 49:534–53 [DOI] [PubMed] [Google Scholar]
- 37.Jhoti H, Rees S, Solari R. High-throughput screening and structure-based approaches to hit discovery: is there a clear winner? Expert Opin Drug Discov 2013; 8:1449–53 [DOI] [PubMed] [Google Scholar]
- 38.Koshland DE., Jr. Enzyme flexibility and enzyme action. J Cell Comp Physiol 1959; 54:245–58 [DOI] [PubMed] [Google Scholar]
- 39.Kumar S, Ma B, Tsai CJ, Sinha N, Nussinov R. Folding and binding cascades: dynamic landscapes and population shifts. Protein Sci 2000; 9:10–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rubin MM, Changeux JP. On the nature of allosteric transitions: implications of non-exclusive ligand binding. J Mol Biol 1966; 21:265–74 [DOI] [PubMed] [Google Scholar]
- 41.Lavecchia A, Di Giovanni C. Virtual screening strategies in drug discovery: a critical review. Curr Med Chem 2013; 20:2839–60 [DOI] [PubMed] [Google Scholar]
- 42.Kinnings SL, Liu N, Tonge PJ, Jackson RM, Xie L, Bourne PE. A machine learning-based method to improve docking scoring functions and its application to drug repurposing. J Chem Inf Model 2011; 51:408–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rodriguez D, Gutierrez-de-Teran H. Computational approaches for ligand discovery and design in class-A G protein-coupled receptors. Curr Pharm Des 2013; 19:2216–36 [DOI] [PubMed] [Google Scholar]
- 44.Martin YC, Kofron JL, Traphagen LM. Do structurally similar molecules have similar biological activity? J Med Chem 2002; 45:4350–8 [DOI] [PubMed] [Google Scholar]
- 45.Favia AD. Theoretical and computational approaches to ligand-based drug discovery. Front Biosci 2011; 16:1276–90 [DOI] [PubMed] [Google Scholar]
- 46.Cheng J, Tegge AN, Baldi P. Machine learning methods for protein structure prediction. IEEE Rev Biomed Eng 2008; 1:41–9 [DOI] [PubMed] [Google Scholar]
- 47.Jabeen A, Ranganathan S. Applications of machine learning in GPCR bioactive ligand discovery. Curr Opin Struct Biol 2019; 55:66–76 [DOI] [PubMed] [Google Scholar]
- 48.Rosenbaum DM, Rasmussen SG, Kobilka BK. The structure and function of G-protein-coupled receptors. Nature 2009; 459:356–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mahoney JP, Sunahara RK. Mechanistic insights into GPCR-G protein interactions. Curr Opin Struct Biol 2016; 41:247–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gurevich VV, Gurevich EV. Molecular mechanisms of GPCR signaling: a structural perspective. Int J Mol Sci 2017; 18:2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang J, Hua T, Liu ZJ. Structural features of activated GPCR signaling complexes. Curr Opin Struct Biol 2020; 63:82–9 [DOI] [PubMed] [Google Scholar]
- 52.Manglik A, Kruse AC. Structural basis for G protein-coupled receptor activation. Biochemistry 2017; 56:5628–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Burg JS, Ingram JR, Venkatakrishnan AJ, Jude KM, Dukkipati A, Feinberg EN, Angelini A, Waghray D, Dror RO, Ploegh HL, Garcia KC. Structural biology. Structural basis for chemokine recognition and activation of a viral G protein-coupled receptor. Science 2015; 347:1113–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dror RO, Green HF, Valant C, Borhani DW, Valcourt JR, Pan AC, Arlow DH, Canals M, Lane JR, Rahmani R, Baell JB, Sexton PM, Christopoulos A, Shaw DE. Structural basis for modulation of a G-protein-coupled receptor by allosteric drugs. Nature 2013; 503:295–9 [DOI] [PubMed] [Google Scholar]
- 55.Guo D, Pan AC, Dror RO, Mocking T, Liu R, Heitman LH, Shaw DE, Ij AP. Molecular basis of ligand dissociation from the adenosine A2A receptor. Mol Pharmacol 2016; 89:485–91 [DOI] [PubMed] [Google Scholar]
- 56.McRobb FM, Negri A, Beuming T, Sherman W. Molecular dynamics techniques for modeling G protein-coupled receptors. Curr Opin Pharmacol 2016; 30:69–75 [DOI] [PubMed] [Google Scholar]
- 57.Ciancetta A, Sabbadin D, Federico S, Spalluto G, Moro S. Advances in computational techniques to study GPCR-Ligand recognition. Trends Pharmacol Sci 2015; 36:878–90 [DOI] [PubMed] [Google Scholar]
- 58.Chan HCS, Xu Y, Tan L, Vogel H, Cheng J, Wu D, Yuan S. Enhancing the signaling of GPCRs via orthosteric ions. ACS Cent Sci 2020; 6:274–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yuan X, Raniolo S, Limongelli V, Xu Y. The molecular mechanism underlying ligand binding to the membrane-embedded site of a G-protein-coupled receptor. J Chem Theory Comput 2018; 14:2761–70 [DOI] [PubMed] [Google Scholar]
- 60.Bueschbell B, Barreto CAV, Preto AJ, Schiedel AC, Moreira IS. A complete assessment of dopamine receptor- ligand interactions through. Comput Meth Mol 2019; 24:1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Salmaso V, Jacobson KA. In silico drug design for purinergic GPCRs: overview on molecular dynamics applied to adenosine and P2Y receptors. Biomolecules 2020; 10:812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang J, Miao Y. Mechanistic insights into specific G protein interactions with adenosine receptors. J Phys Chem B 2019; 123:6462–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schneider G. Automating drug discovery. Nat Rev Drug Discov 2018; 17:97–113 [DOI] [PubMed] [Google Scholar]
- 64.Plante A, Shore DM, Morra G, Khelashvili G, Weinstein H. A machine learning approach for the discovery of ligand-specific functional mechanisms of GPCRs. Molecules 2019; 24:2097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Schonenbach NS, Hussain S, O'Malley MA. Structure and function of G protein-coupled receptor oligomers: implications for drug discovery. Wiley Interdiscip Rev Nanomed Nanobiotechnol 2015; 7:408–27 [DOI] [PubMed] [Google Scholar]
- 66.Topiol S. Current and future challenges in GPCR drug discovery. Methods Mol Biol 2018; 1705:1–21 [DOI] [PubMed] [Google Scholar]
- 67.Latorraca NR, Venkatakrishnan AJ, Dror RO. GPCR dynamics: structures in motion. Chem Rev 2017; 117:139–55 [DOI] [PubMed] [Google Scholar]
- 68.Borodovsky A, Barbon CM, Wang Y, Ye M, Prickett L, Chandra D, Shaw J, Deng N, Sachsenmeier K, Clarke JD, Linghu B, Brown GA, Brown J, Congreve M, Cheng RK, Dore AS, Hurrell E, Shao W, Woessner R, Reimer C, Drew L, Fawell S, Schuller AG, Mele DA. Small molecule AZD4635 inhibitor of A2AR signaling rescues immune cell function including CD103(+) dendritic cells enhancing anti-tumor immunity. J Immunother Cancer 2020; 8:e000417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Christ CD, Mark AE, van Gunsteren WF. Basic ingredients of free energy calculations: a review. J Comput Chem 2010; 31:1569–82 [DOI] [PubMed] [Google Scholar]
- 70.Reddy MR, Reddy CR, Rathore RS, Erion MD, Aparoy P, Reddy RN, Reddanna P. Free energy calculations to estimate ligand-binding affinities in structure-based drug design. Curr Pharm Des 2014; 20:3323–37 [DOI] [PubMed] [Google Scholar]
- 71.Pires DEV, Veloso WNP, Myung Y, Rodrigues CHM, Silk M, Rezende PM, Silva F, Xavier JS, Velloso JPL, da Silveira CH, Ascher DB. EasyVS: a user-friendly web-based tool for molecule library selection and structure-based virtual screening. Bioinformatics 2020; 36:4200–2 [DOI] [PubMed] [Google Scholar]
- 72.de Vries SJ, Rey J, Schindler CEM, Zacharias M, Tuffery P. The pepATTRACT web server for blind, large-scale peptide-protein docking. Nucleic Acids Res 2017; 45:W361–W64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Fenollosa C, Oton M, Andrio P, Cortes J, Orozco M, Goni JR. SEABED: small molecule activity scanner weB servicE baseD. Bioinformatics 2015; 31:773–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Weng G, Wang E, Wang Z, Liu H, Zhu F, Li D, Hou T. HawkDock: a web server to predict and analyze the protein-protein complex based on computational docking and MM/GBSA. Nucleic Acids Res 2019; 47:W322–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Labbe CM, Pencheva T, Jereva D, Desvillechabrol D, Becot J, Villoutreix BO, Pajeva I, Miteva MA. AMMOS2: a web server for protein-ligand-water complexes refinement via molecular mechanics. Nucleic Acids Res 2017; 45:W350–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Labbe CM, Rey J, Lagorce D, Vavrusa M, Becot J, Sperandio O, Villoutreix BO, Tuffery P, Miteva MA. MTiOpenScreen: a web server for structure-based virtual screening. Nucleic Acids Res 2015; 43:W448–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Santos KB, Guedes IA, Karl ALM, Dardenne LE. Highly flexible ligand docking: benchmarking of the DockThor program on the LEADS-PEP Protein-Peptide data set. J Chem Inf Model 2020; 60:667–83 [DOI] [PubMed] [Google Scholar]
- 78.Firestein S. How the olfactory system makes sense of scents. Nature 2001; 413:211–8 [DOI] [PubMed] [Google Scholar]
- 79.Bavan S, Sherman B, Luetje CW, Abaffy T. Discovery of novel ligands for mouse olfactory receptor MOR42-3 using an in silico screening approach and in vitro validation. PLoS One 2014; 9:e92064.[ 10.1371/journal.pone.0092064] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yuan S, Dahoun T, Brugarolas M, Pick H, Filipek S, Vogel H. Computational modeling of the olfactory receptor Olfr73 suggests a molecular basis for low potency of olfactory receptor-activating compounds. Commun Biol 2019; 2:141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wisler JW, Xiao KH, Thomsen ARB, Lefkowitz RJ. Recent developments in biased agonism. Curr Opin Cell Biol 2014; 27:18–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wisler JW, Rockman HA, Lefkowitz RJ. Biased G protein-coupled receptor signaling changing the paradigm of drug discovery. Circulation 2018; 137:2315–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Bermudez M, Nguyen TN, Omieczynski C, Wolber G. Strategies for the discovery of biased GPCR ligands. Drug Discov Today 2019; 24:1031–7 [DOI] [PubMed] [Google Scholar]
- 84.Costa-Neto CM, Parreiras ESLT, Bouvier M. A pluridimensional view of biased agonism. Mol Pharmacol 2016; 90:587–95 [DOI] [PubMed] [Google Scholar]
- 85.Weiss DR, Ahn S, Sassano MF, Kleist A, Zhu X, Strachan R, Roth BL, Lefkowitz RJ, Shoichet BK. Conformation guides molecular efficacy in docking screens of activated beta-2 adrenergic G protein coupled receptor. ACS Chem Biol 2013; 8:1018–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Brogi S, Tafi A, Desaubry L, Nebigil CG. Discovery of GPCR ligands for probing signal transduction pathways. Front Pharmacol 2014; 5:255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Mannel B, Jaiteh M, Zeifman A, Randakova A, Moller D, Hubner H, Gmeiner P, Carlsson J. Structure-Guided screening for functionally selective D2 dopamine receptor ligands from a virtual chemical library. ACS Chem Biol 2017; 12:2652–61 [DOI] [PubMed] [Google Scholar]
- 88.McCorvy JD, Butler KV, Kelly B, Rechsteiner K, Karpiak J, Betz RM, Kormos BL, Shoichet BK, Dror RO, Jin J, Roth BL. Structure-inspired design of beta-arrestin-biased ligands for aminergic GPCRs. Nat Chem Biol 2018; 14:126–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Patel N, Huang XP, Grandner JM, Johansson LC, Stauch B, McCorvy JD, Liu Y, Roth B, Katritch V. Structure-based discovery of potent and selective melatonin receptor agonists. Elife 2020; 9:e53779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Marti-Solano M, Iglesias A, de Fabritiis G, Sanz F, Brea J, Loza MI, Pastor M, Selent J. Detection of new biased agonists for the serotonin 5-HT2A receptor: modeling and experimental validation. Mol Pharmacol 2015; 87:740–6 [DOI] [PubMed] [Google Scholar]
- 91.Fischer T, Gazzola S, Riedl R. Approaching target selectivity by de novo drug design. Expert Opin Drug Discov 2019; 14:791–803 [DOI] [PubMed] [Google Scholar]
- 92.Schneider G, Clark DE. Automated de novo drug design: are we nearly there yet? Angew Chem Int Ed Engl 2019; 58:10792–803 [DOI] [PubMed] [Google Scholar]
- 93.Li Y, Sun Y, Song Y, Dai D, Zhao Z, Zhang Q, Zhong W, Hu LA, Ma Y, Li X, Wang R. Fragment-based computational method for designing GPCR ligands. J Chem Inf Model 2020; 60:4339–49 [DOI] [PubMed] [Google Scholar]
- 94.Basith S, Cui M, Macalino SJY, Park J, Clavio NAB, Kang S, Choi S. Exploring G protein-coupled receptors (GPCRs) ligand space via cheminformatics approaches: impact on rational drug design. Front Pharmacol 2018; 9:128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Dedic N, Jones PG, Hopkins SC, Lew R, Shao L, Campbell JE, Spear KL, Large TH, Campbell UC, Hanania T, Leahy E, Koblan KS. SEP-363856, a novel psychotropic agent with a unique, non-D2 receptor mechanism of action. J Pharmacol Exp Ther 2019; 371:1–14 [DOI] [PubMed] [Google Scholar]
- 96.Kawai K, Nagata N, Takahashi Y. De novo design of drug-like molecules by a fragment-based molecular evolutionary approach. J Chem Inf Model 2014; 54:49–56 [DOI] [PubMed] [Google Scholar]
- 97.Reutlinger M, Rodrigues T, Schneider P, Schneider G. Multi-objective molecular de novo design by adaptive fragment prioritization. Angew Chem Int Ed Engl 2014; 53:4244–8 [DOI] [PubMed] [Google Scholar]
- 98.Fedorov DG, Nagata T, Kitaura K. Exploring chemistry with the fragment molecular orbital method. Phys Chem Chem Phys 2012; 14:7562–77 [DOI] [PubMed] [Google Scholar]
- 99.Chudyk EI, Sarrat L, Aldeghi M, Fedorov DG, Bodkin MJ, James T, Southey M, Robinson R, Morao I, Heifetz A. Exploring GPCR-Ligand interactions with the fragment molecular orbital (FMO) method. Methods Mol Biol 2018; 1705:179–95 [DOI] [PubMed] [Google Scholar]
- 100.Heifetz A, Chudyk EI, Gleave L, Aldeghi M, Cherezov V, Fedorov DG, Biggin PC, Bodkin MJ. The fragment molecular orbital method reveals new insight into the chemical nature of GPCR-ligand interactions. J Chem Inf Model 2016; 56:159–72 [DOI] [PubMed] [Google Scholar]
- 101.Heifetz A, James T, Southey M, Morao I, Aldeghi M, Sarrat L, Fedorov DG, Bodkin MJ, Townsend-Nicholson A. Characterising GPCR-ligand interactions using a fragment molecular orbital-based approach. Curr Opin Struct Biol 2019; 55:85–92 [DOI] [PubMed] [Google Scholar]
- 102.Chen H, Engkvist O, Wang Y, Olivecrona M, Blaschke T. The rise of deep learning in drug discovery. Drug Discov Today 2018; 23:1241–50 [DOI] [PubMed] [Google Scholar]
- 103.Cherkasov A, Muratov EN, Fourches D, Varnek A, Baskin II, Cronin M, Dearden J, Gramatica P, Martin YC, Todeschini R, Consonni V, Kuz'min VE, Cramer R, Benigni R, Yang C, Rathman J, Terfloth L, Gasteiger J, Richard A, Tropsha A. QSAR modeling: where have you been? Where are you going to? J Med Chem 2014; 57:4977–5010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Rogers D, Hahn M. Extended-connectivity fingerprints. J Chem Inf Model 2010; 50:742–54 [DOI] [PubMed] [Google Scholar]
- 105.Jaeger S, Fulle S, Turk S. Mol2vec: unsupervised machine learning approach with chemical intuition. J Chem Inf Model 2018; 58:27–35 [DOI] [PubMed] [Google Scholar]
- 106.Kadurin A, Nikolenko S, Khrabrov K, Aliper A, Zhavoronkov A. druGAN: an advanced generative adversarial autoencoder model for de novo generation of new molecules with desired molecular properties in silico. Mol Pharm 2017; 14:3098–104 [DOI] [PubMed] [Google Scholar]
- 107.Liu X, Ye K, van Vlijmen HWT, AI, van Westen GJP. An exploration strategy improves the diversity of de novo ligands using deep reinforcement learning: a case for the adenosine A2A receptor. J Cheminform 2019; 11:35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Rataj K, Kelemen AA, Brea J, Loza MI, Bojarski AJ, Keseru GM. Fingerprint-based machine learning approach to identify potent and selective 5-HT2BR ligands. Molecules 2018; 23:1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Crasto C, Marenco L, Miller P, Shepherd G. Olfactory receptor database: a metadata-driven automated population from sources of gene and protein sequences. Nucleic Acids Res 2002; 30:354–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Bushdid C, de March CA, Fiorucci S, Matsunami H, Golebiowski J. Agonists of G-protein-coupled odorant receptors are predicted from chemical features. J Phys Chem Lett 2018; 9:2235–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Priest BT, Bell IM, Garcia ML. Role of hERG potassium channel assays in drug development. Channels 2008; 2:87–93 [DOI] [PubMed] [Google Scholar]
- 112.Ryu JY, Lee MY, Lee JH, Lee BH, Oh KS. DeepHIT: a deep learning framework for prediction of hERG-induced cardiotoxicity. Bioinformatics 2020; 36:3049–55 [DOI] [PubMed] [Google Scholar]
- 113.Villoutreix BO, Taboureau O. Computational investigations of hERG channel blockers: new insights and current predictive models. Adv Drug Deliv Rev 2015; 86:72–82 [DOI] [PubMed] [Google Scholar]
- 114.Wang S, Li Y, Xu L, Li D, Hou T. Recent developments in computational prediction of HERG blockage. Curr Top Med Chem 2013; 13:1317–26 [DOI] [PubMed] [Google Scholar]
- 115.Taboureau O, Jorgensen FS. In silico predictions of hERG channel blockers in drug discovery: from ligand-based and target-based approaches to systems chemical biology. Comb Chem High Throughput Screen 2011; 14:375–87 [DOI] [PubMed] [Google Scholar]
- 116.Negami T, Araki M, Okuno Y, Terada T. Calculation of absolute binding free energies between the hERG channel and structurally diverse drugs. Sci Rep 2019; 9:16586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Du-Cuny L, Chen L, Zhang S. A critical assessment of combined ligand- and structure-based approaches to HERG channel blocker modeling. J Chem Inf Model 2011; 51:2948–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Braga RC, Alves VM, Silva MF, Muratov E, Fourches D, Liao LM, Tropsha A, Andrade CH. Pred-hERG: a novel web-accessible computational tool for predicting cardiac toxicity. Mol Inform 2015; 34:698–701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Chemi G, Gemma S, Campiani G, Brogi S, Butini S, Brindisi M. Computational tool for fast in silico evaluation of hERG K(+) channel affinity. Front Chem 2017; 5:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Zhang YM, Zhao JN, Wang YC, Fan YR, Zhu L, Yang Y, Chen XY, Lu T, Chen YD, Liu HC. Prediction of hERG K plus channel blockage using deep neural networks. Chem Biol Drug Des 2019; 94:1973–85 [DOI] [PubMed] [Google Scholar]
- 121.Cai C, Guo P, Zhou Y, Zhou J, Wang Q, Zhang F, Fang J, Cheng F. Deep learning-based prediction of drug-induced cardiotoxicity. J Chem Inf Model 2019; 59:1073–84 [DOI] [PMC free article] [PubMed] [Google Scholar]