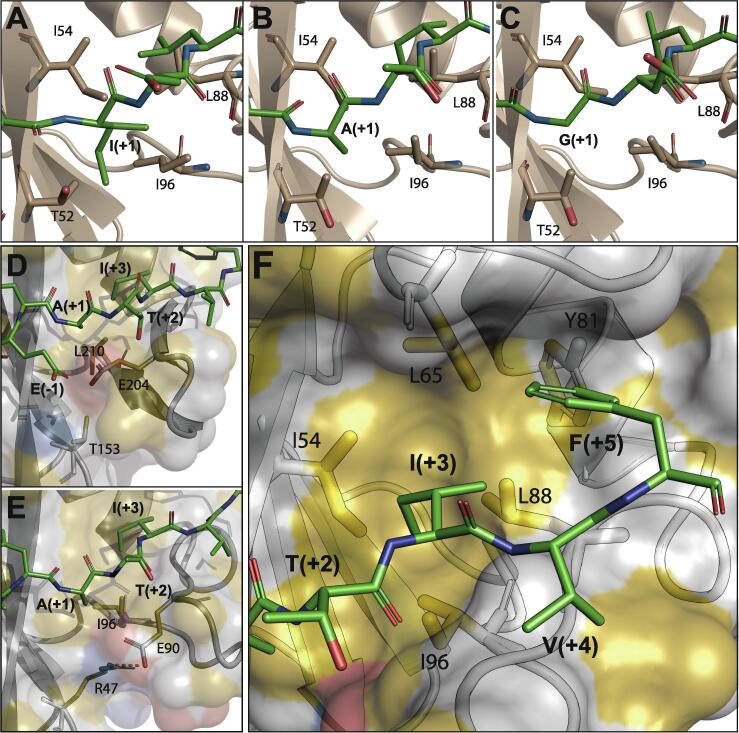
Fig. 2.
Recognition of phosphopeptide sidechains at positions pY+1, pY+3 and pY+5. (A, B, C) Interactions between the hydrophobic cluster of N-SH2, formed by I54, L88 and I96, and the pY+1 residue of IRS1-pY1172, ITSM and ITIM, respectively. (D) Same region of the protein as in panels A–C, but for the C-SH2–ITSM complex. E204 in C-SH2 cannot form a salt-bridge with T153 (for which the corresponding residue in N-SH2 is R47) and thus shields L210 from interacting with the peptide residue pY+1. (E) Same as panel B (N-SH2–ITIM complex), highlighting the salt-bridge between E90 and R47, which exposes I96 to the ITSM residue pY+1. (F) The pY+5 binding site of N-SH2 bound to ITSM consists of a pentagonal arrangement of five hydrophobic residues (I54, L65, Y81, L88 & I96), which also contact the side-chain of the pY+3 residue.