FIGURE 4.
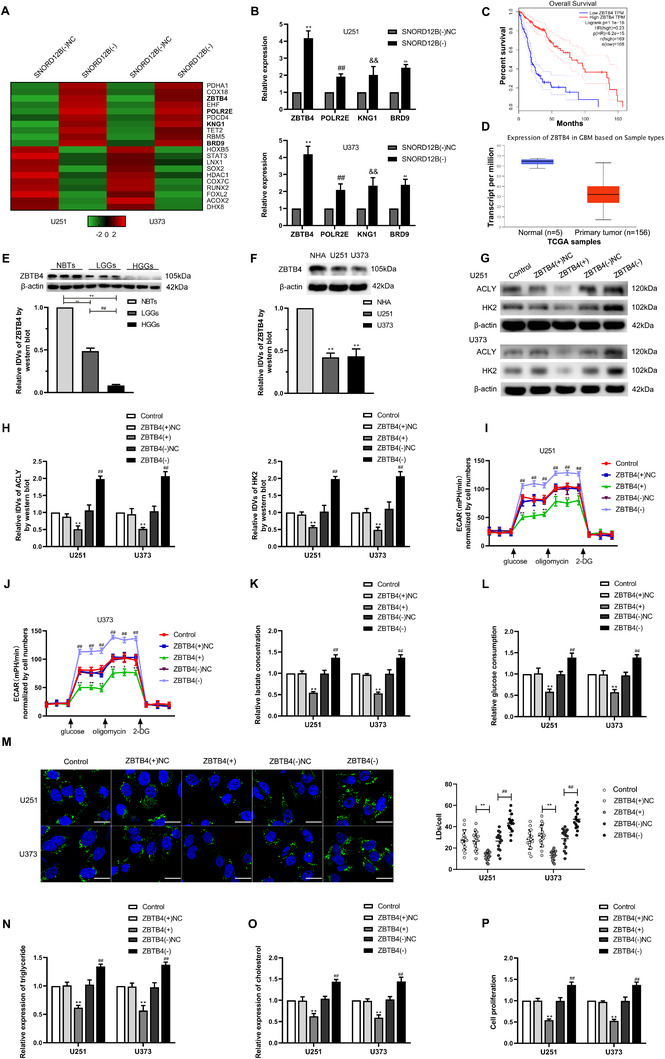
ZBTB4 was downregulated in GBM tissues and cells, and overexpression of ZBTB4 inhibited GBM cells glycolipid metabolism and proliferation. (A) Whole human genome microarray was performed to detect the gene profile when SNORD12B was knockdown. (B) qRT‐PCR was performed to validate the selected molecules. **/##/&&/ΔΔ p < .01 versus SNORD(−)NC group. (C) Effect of ZBTB4 expression level on glioma patient survival time from TCGA database. (D) Expression of ZBTB4 in NBTs and glioma from TCGA samples. (E) ZBTB4 protein levels were analyzed in NBTs, LGGs, and HGGs by Western blot. ** p < .01 versus NBTs group; ## p < .01 versus LGGs group. (F) ZBTB4 protein levels in NHA, U251, and U373 cells were detected via Western blot. ** p < .01 versus NHA group. (G and H) Regulation of HK2 and ACLY expression by ZBTB4 was analyzed via Western blot. (I and J) Effect of ZBTB4 on glycolysis and glycolytic capacity in U251 and U373 cells was measured via ECAR. (K and L) Lactate production and glucose uptake were measured in U251 and U373 cells after ZBTB4 knockdown or overexpression. * p < .05 versus ZBTB4(+)NC group; ** p < .01 versus ZBTB4(+)NC group; ## p < .01 versus ZBTB4(−)NC group. (M) Representative confocal fluorescence imaging of LDs stained by BODIPY 493/503 (green) in U251 and U373 cells after ZBTB4 knockdown or overexpression. Nucleus (blue) was stained by DAPI. Scale bars = 20 μm. Data are presented as the mean ± SD (n = 15, each group). ** p < .01 versus ZBTB4(+)NC group; ## p < .01 versus ZBTB4(−)NC group. (N and O) Intracellular triglyceride and cholesterol expression levels were measured to evaluate the effect of ZBTB4 on lipogenesis. (P) Effect of ZBTB4 on proliferation was analyzed via CCK‐8 assay. ** p < .01 versus ZBTB4(+)NC group; ## p < .01 versus ZBTB4(−)NC group. Except for specially noted, data are presented as the mean ± SD of three independent experiments per group. Statistical analysis was by one‐way ANOVA method
