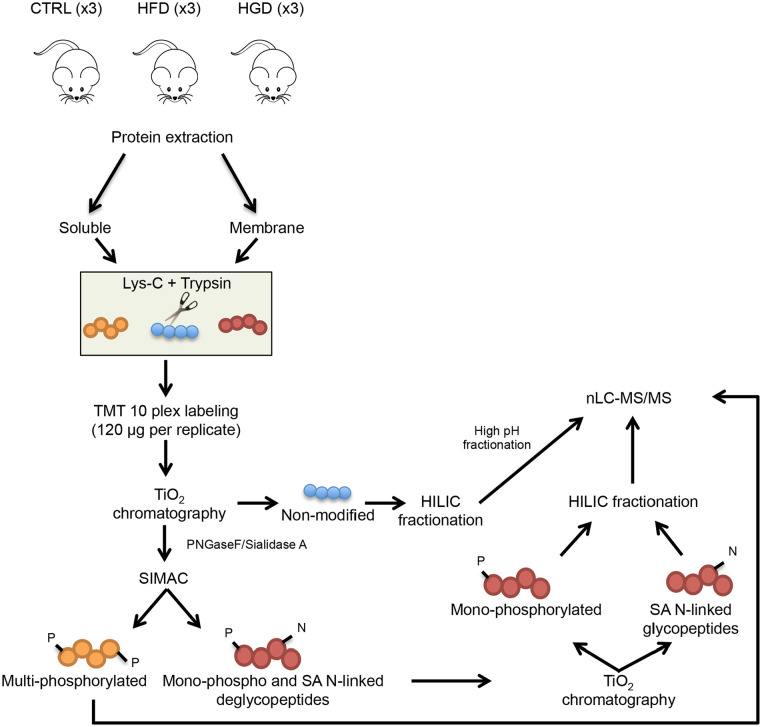
Figure 1.
Workflow of quantitative mass spectrometry-based proteomic approach. After mice sacrifice, brains were collected from 3 mice fed with a standard diet (SD), 3 mice fed with high-fat diet (HFD) and 3 mice fed with high-glycaemic diet (HGD) and proteins were extracted. Soluble and membrane protein fractions were separated by ultracentrifugation followed by reduction, alkylation and digestion. Both fractions were TMT labelled and combined. Multiple enrichment steps using TiO2 and sequential elution from IMAC (SIMAC) beads resulted in 3 proteomic pools: non-modified peptides, phosphorylated peptides and formerly sialylated (SA) N-linked glycopeptides. Complexity of samples was reduced using HILIC fractionation for non-modified, mono-phosphorylated and SA N-linked peptides. Moreover, non-modified peptides were also fractionated by high pH and each fraction was run by nLC-MS/MS.