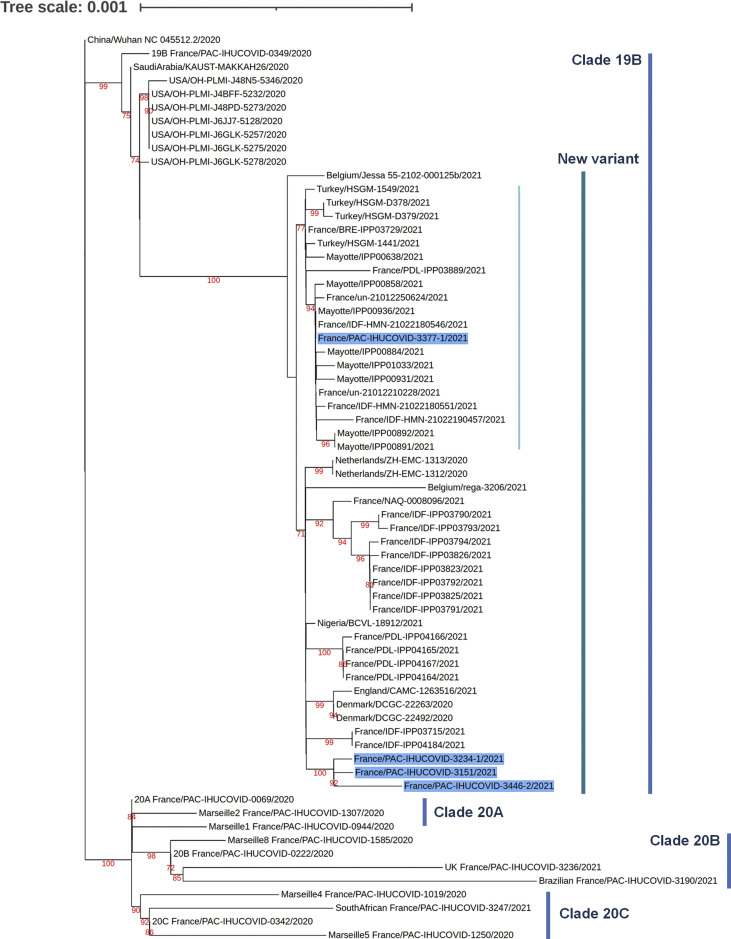
Fig. 2.
Phylogenetic analysis of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) full-length genomes of the new variant recovered from the IHU Méditerranée Infection sequence database and from the GISAID sequence database. The SARS-CoV-2 phylogenetic tree is based on the full-length SARS-CoV-2 genomes. The SARS-CoV-2 sequences obtained by next-generation sequencing from four patients are indicated by a bold font and a dark blue background. The sequences with the highest BLAST scores recovered from the NBCI GenBank nucleotide sequence databases (http://www.ncbi.nlm.nih.gov/nucleotide/) including the 42 that are clustered with these four query sequences, were incorporated in the phylogeny reconstruction. Additional sequences include the genome of the Wuhan-Hu-1 isolate and genomes obtained in our institute and classified as predominant variants. The 67 genomes of SARS-CoV-2 were aligned using MAFFT version 7 with default parameters [10]. Phylogenetic tree was performed by using iQ-TREE under the GTR model and visualized with iTOL [[9], [11]]. GISAID accession numbers: IHUCOVID-3234: EPI_ISL_1097023; IHUCOVID-3151: EPI_ISL_1097024; IHUCOVID-3377: EPI_ISL_1201045; IHUCOVID-3446: EPI_ISL_1201046.