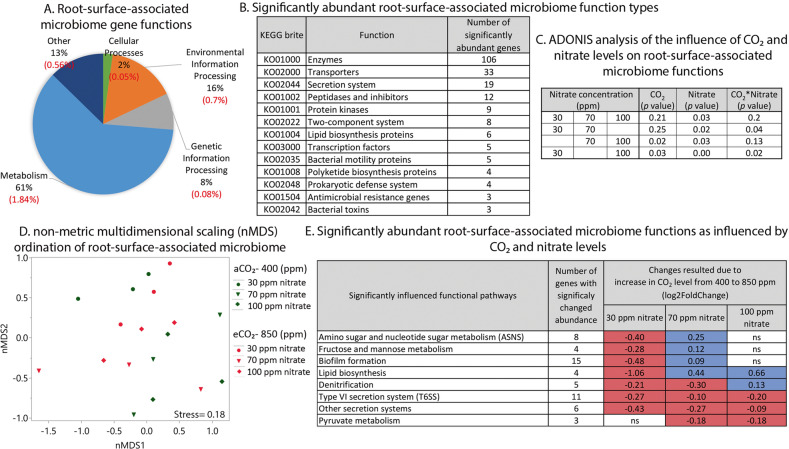
Fig. 4. Changes in root microbiome functions (KEGG) of 6-weeks-old wheat as influenced by nitrate and CO2 levels.
Changes in microbiome gene abundance were calculated using DESeq2 with cutoff < 0.2 of FDR-adjusted P value. a Pie chart of all functional genes in wheat root microbiome. Significantly changed pathways are marked in red. In parentheses is the percentage of that pathway out of whole metagenome which was significantly changed as a function of nitrate and CO2 levels. b Significantly abundant root-surface-associated microbiome function types. c ADONIS analysis of summarized effect of CO2 and different combinations of nitrate supply on bacterial community functional genes. d nMDS ordination plot showing clustering patterns of root-surface-associated microbiome functional genes as influenced by nitrate and CO2 levels. Data matrix was transformed using normalized count transformation and batch effect removal with the DESeq2 package, and then ordination was generated using Euclidean dissimilarity, n = 3. e Significantly abundant root-surface-associated microbiome functional gene pathways with increase in CO2 level. Blue indicates positive and red indicates negative log2 fold change in gene abundance between 850 and 400 ppm CO2. ns not significant.