Fig. 1. This example shows that a loss of a terminal segment of a chromosome arm, as detected by CMA or WGS (in red), can be caused by four different structural rearrangements.
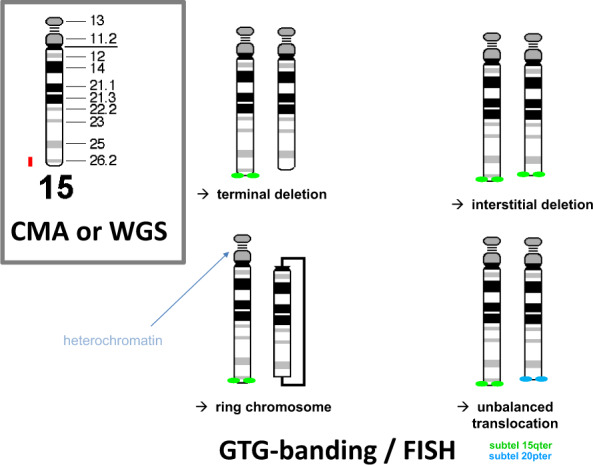
Current WGS methods based on short-read, paired-end sequencing are not suited to discriminate between these possible rearrangements because breakpoints that are located within repetitive DNA sequences are not recognized [4]. In contrast, these rearrangements can be discriminated by microscopy, using karyotyping and/or FISH. The simplest structural rearrangement is a terminal deletion although an interstitial deletion, a ring chromosome, or a derivative chromosome representing an unbalanced translocation involving chromosome 15 and the very terminal end of another chromosome arm (here 20p) can underlie such a terminal loss. As emphasized in current guidelines for cytogenomic investigation [7], identification of the underlying rearrangement is essential for determining the recurrence risk for the parents of the patient, for genetic counseling and for the identification of family members who are at an increased risk of having imbalanced progeny.
