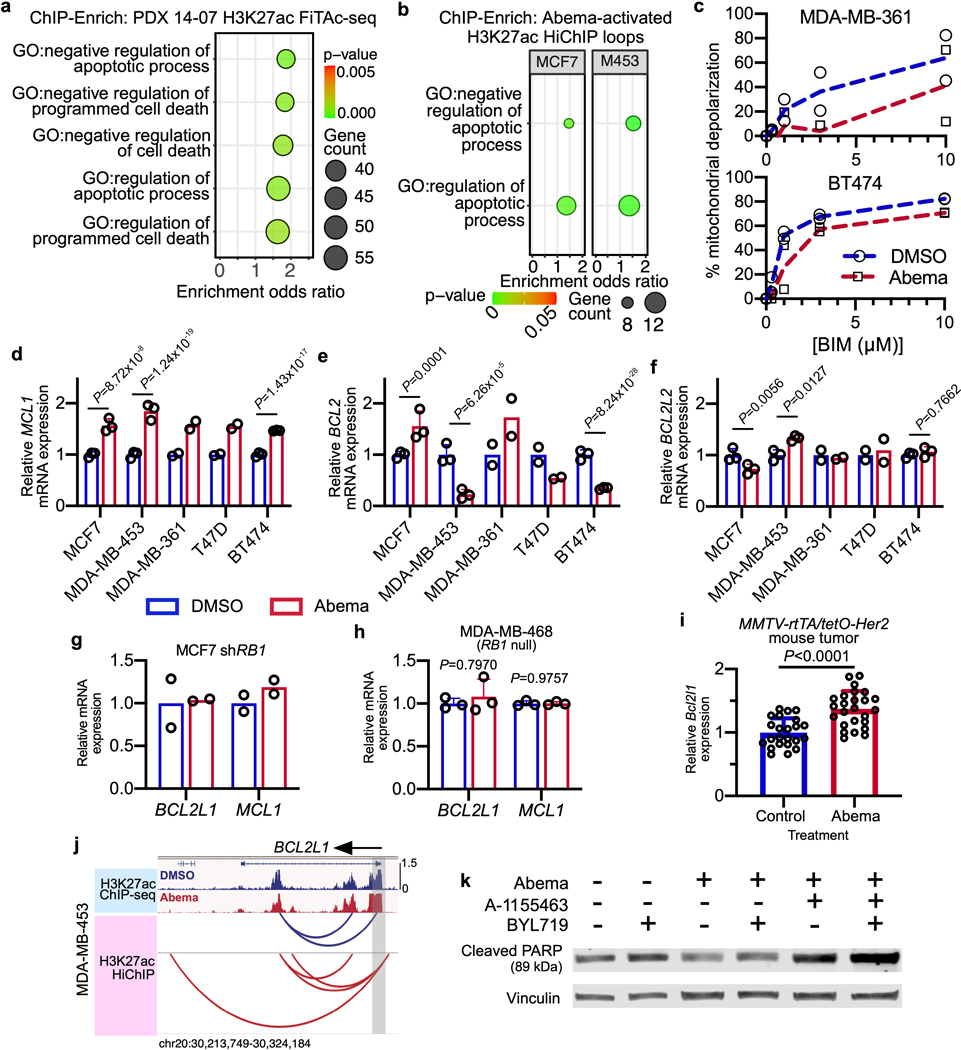
Extended Data Fig. 4. CDK4/6 inhibition promotes apoptotic evasion via BCL2L1.
a,b, ChIP-Enrich analysis of regions gaining H3K27ac after 7 days of abemaciclib treatment of PDX 14–07 tumors (21–28 days of treatment) (a), or of non-TSS ends of genomic loops (measured by HiChIP) detected only in abemaciclib-treated MCF7 and MDA-MB-453 cells (b).
c, Dose-response curves showing percent mitochondrial depolarization in MDA-MB-361 and BT474 treated with DMSO or abemaciclib for 7 days and after exposure to Bim peptide (dynamic BH3 profiling; n=2 technical replicates, data are representative of two independent experiment).
d-f, Relative RNA-seq normalized reads of MCL1 (d), BCL2 (e), and BCL2L2 (f) in human breast cancer cell lines treated with DMSO or abemaciclib, measured by RNA-seq (MDA-MB-361 and T47D: n=2; MCF7, MDA-MB-453, and BT474: n=3 independent cultures). Mean ± s.d. are shown. DESeq2 was used to determine adjusted P-values.
g,h, Relative RNA-seq normalized reads of BCL2L1 and MCL1 in MCF7 shRB1 cells (g; n=2 independent cultures) and MDA-MB-468 (h, RB1 null; n=3 independent cultures) treated with DMSO or abemaciclib. Data are presented as mean ± s.d. DESeq2 was used to calculate adjusted P-values.
i, Relative Bcl2l1 expression by transcriptomic profiling of MMTV-rtTA/tetO-Her2 tumors treated with vehicle (n=23) or abemaciclib (n=25) for 12 days. Mean ± s.d. of normalized reads are shown. P-values were determined using a two-tailed unpaired t-test.
j, H3K27ac ChIP-seq tracks and co-localized H3K27ac-decorated genomic loops (identified by HiChIP) adjacent to BCL2L1 in MDA-MB-453 cells treated with DMSO or abemaciclib for 7 days. Gene promoters are highlighted in grey.
k, Western blot showing cleaved PARP in MCF7 cells treated with DMSO or abemaciclib (500 nM) for 3 days, followed by treatment with A1155463 (1 μM) for 24 hours and BYL719 (1 μM) for 8 hours as indicated. Representative blots of two independent experiments in MCF7. Western blots are cropped; uncropped blot images for the experiments in this figure are shown in Source Data Extended Data Fig. 4.