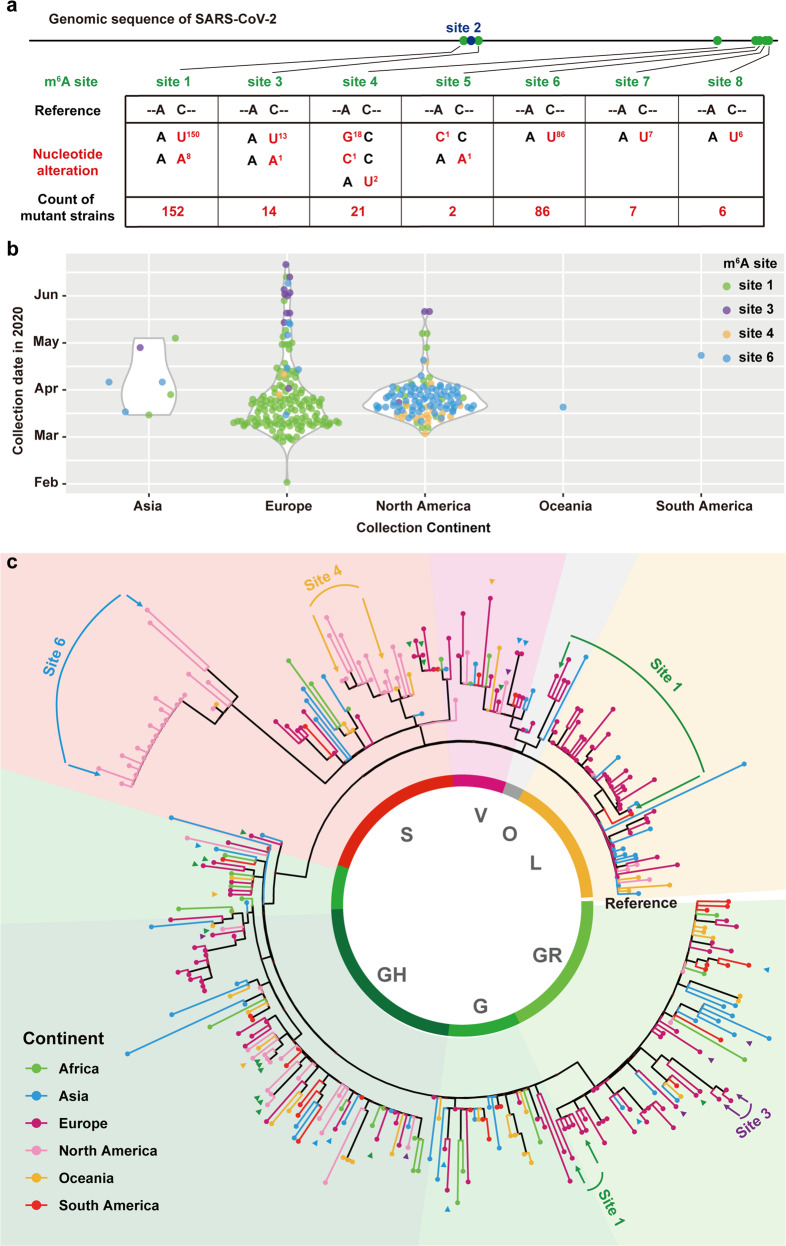
Fig. 2. m6A substitution among diverse SARS-CoV-2 isolates.
a Sequence comparison is performed by python with Biopython package. The EPI_ISL_424359 (blue) is set as the reference sequence. The contemporary SARS-CoV-2 strains used in this study are labeled in black and red. Variations are presented in accordance with their locations in viral genome. b The sample distribution of 289 strains with mutations at m6A modification site 1, 3, 4 and 6, each point represents one sample and plotted by R with ggplot2 package. Most of the mutant strains were found distributed in Europe and North America. c The maximum likelihood phylogenetic tree estimated using IQ-TREE. The final result is visualized by ggtree. Conserved nucleotide changes are inferred using the EPI_ISL_424359 strain as the reference strain. Virus sequences selected from 7 GISAID clade (L, S, G, GH, GR, O, V) and the continent information is shown in different colors. The positions of mutant strains of m6A sites are indicated with arrows colored by green (Site 1), purple (Site 3), yellow (Site 4) and blue (Site 6). The details of the tree are shown in Supplementary information, Table S4.