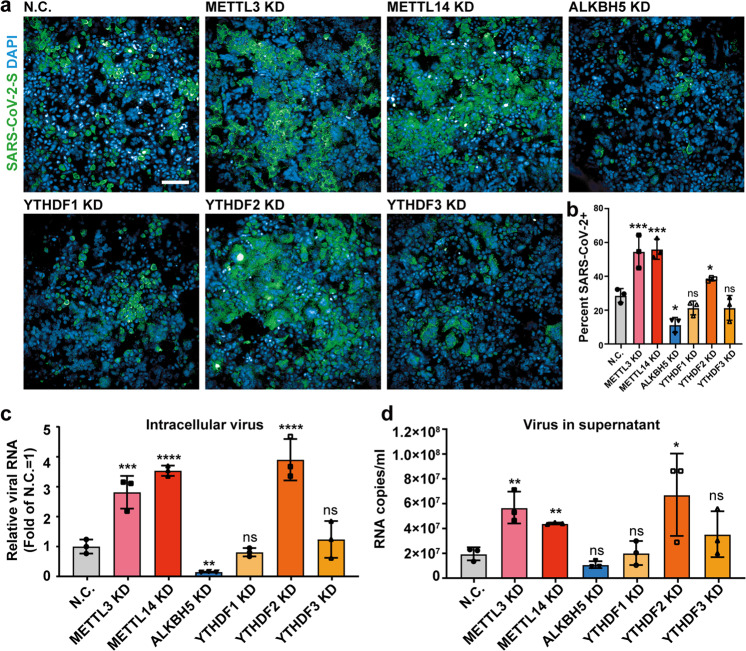
Fig. 4. m6A inhibits SARS-CoV-2 replication.
a Immunofluorescence images of SARS-CoV-2-infected Huh7 cells after m6A methyltransferases (METTL3/14), demethylase (ALKBH5) and reader proteins (YTHDF1, YTHDF2 and YTHDF3) were knocked down, respectively. Huh7 cells were treated with siRNAs targeting METTL3/14, and ALKBH5, meanwhile a non-targeting siRNA as the negative control and then were infected by SARS-CoV-2 at an MOI of 0.05 for 72 h. Specific antibodies recognizing the SARS-CoV-2 S protein (green) were used; DNA was stained with DAPI (blue). Scale bar, 20 µm. b Percentage of viral S protein-positive cells as treated in a. n = 3. Statistical significance of the difference was determined by Student’s unpaired t-test. *P < 0.05; ***P < 0.001; ns, non-significant. c Relative SARS-CoV-2 RNA copies in siRNA-treated cells quantified by qRT-PCR at 72 hpi. RNA was normalized with GAPDH. n = 3. All data are the means ± SD of the indicated number of replicates. Statistical significance of the difference was determined by Student’s unpaired t-test. **P < 0.01; ***P < 0.001; ****P < 0.0001; ns, non-significant. d The released SARS-CoV-2 RNA in siRNA-treated supernatants was quantified by qRT-PCR at 72 hpi. n = 3. All data are the means ± SD of the indicated number of replicates. Statistical significance of the difference was determined by Student’s unpaired t-test. *P < 0.05; **P < 0.01; ns, non-significant.