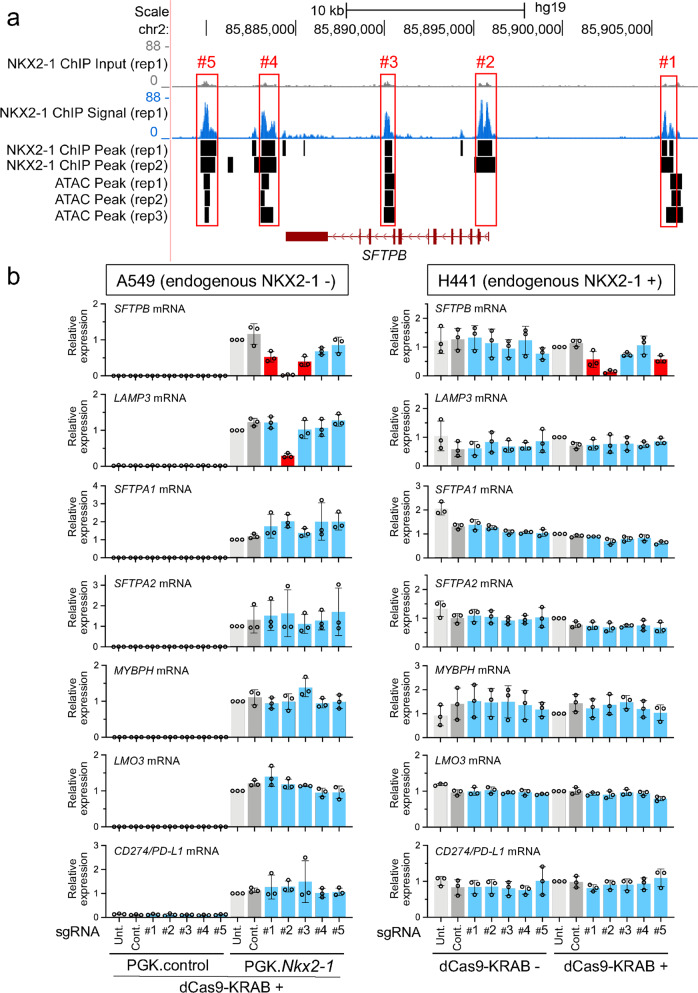
Fig. 2. NKX2-1-binding sites located at the upstream and first intronic regions are commonly required for the expression of SFTPB induced by NKX2-1.
a Shown are ATAC-seq and ChIP-seq data indicating that NKX2-1 binds to upstream (#1), first (#2), ninth intronic (#3), proximal (#4), and distal downstream (#5) regions of human SFTPB locus. ChIP-seq was previously performed using an A549 lung epithelial cell line stably expressing NKX2-127,30. ATAC-seq was obtained from the ENCODE database37. NKX2-1-binding sites are shown as peaks obtained from bam files. Significant peaks compared to input peaks are shown as side bars obtained from bed files. b Shown is the gene expression of NKX2-1 downstream target genes affected by CRISPRi from biologically independent triplicates, in which synthetic sgRNAs targeting NKX2-1-binding sites (#) located at the regions described in (a) are transiently transfected into A549 cells expressing dCas9-KRAB with or without NKX2-1 (PGK.Nkx2-1 or PGK vector control), or H441 cells with or without dCas9-KRAB. The sgRNA targeting NKX2-1-binding sites located at the upstream (#1) and first intronic (#2) regions significantly suppressed the expression of SFTPB in both A549 and H441 cells. The sgRNA targeting NKX2-1-binding sites located at the first intronic (#2) region also significantly suppressed the expression of LAMP3 induced by NKX2-1 in A549 cells. Of note, endogenous NKX2-1 is expressed in H441 cells but not in A549 cells (see Supplementary Fig. 1). Results are expressed as the mean ± SD of the triplicates for each group. Only P < 0.05 and more than two-fold suppression are considered significant CRISPRi-mediated suppression (highlighted in red). Unt., untransfected. Cont., non-targeted control sgRNA.