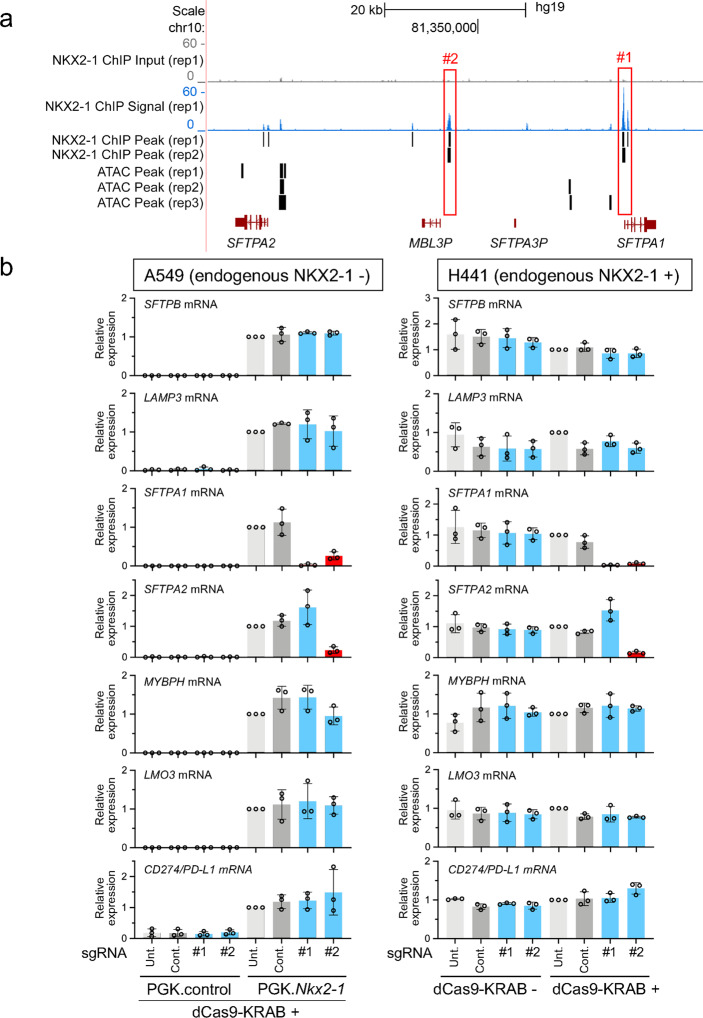
Fig. 5. NKX2-1-binding sites located at two different regions between SFTPA1 and SFTPA2 function distinctively for the expression of SFTPA1 and SFTPA2 in both A549 and H441 cells.
a Shown are ATAC-seq and ChIP-seq data indicating that NKX2-1 binds to an intergenic (#1 and #2) region of human SFTPA1 and SFTPA2. Significant peaks are described as in Fig. 2a. MBL3P and SFTPA3P are pseudogenes. b Shown is gene expression of NKX2-1 downstream target genes affected by CRISPRi as described in Fig. 2b. The sgRNA targeting an NKX2-1-binding site located at the proximal upstream (#1) region of SFTPA1 significantly suppressed the NKX2-1-induced expression of SFTPA1 but not SFTPA2, while the one targeting an NKX2-1-binding site located at the distal upstream (#2) region of SFTPA1 and SFTPA2 significantly suppressed the NKX2-1-induced expression of both SFTPA1 and SFTPA2. Results are expressed as the mean ± SD of the triplicates for each group. Only P < 0.05 and more than twofold suppression are considered significant CRISPRi-mediated suppression (highlighted in red). Unt. untransfected, Cont. non-targeted control sgRNA.