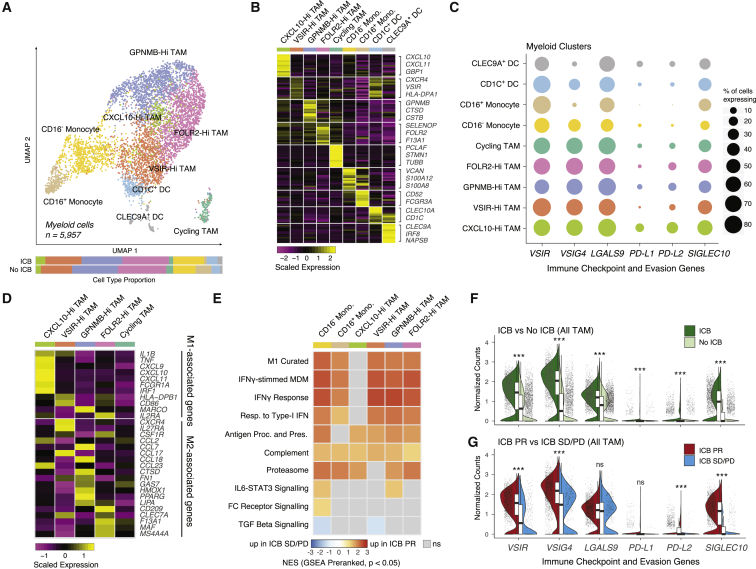
Figure 3.
Tumor-associated macrophages shift toward inflammation during checkpoint blockade
(A) UMAP of myeloid cells captured across all lesions, colored and labeled by cell type. Bar plots show cell-type proportions grouped by ICB treatment history.
(B) Heatmap of scaled normalized expression for cell-type-defining genes as determined by two-sided Wilcoxon rank-sum test with Bonferroni FDR correction (q < 0.01).
(C) Dotplot showing percentage of cells in each cell type expressing immune checkpoint and evasion genes.
(D) Heatmap of scaled normalized expression for curated M1- and M2-associated genes within TAM subtypes.
(E) Heatmap of normalized enrichment scores (NES) for gene sets significantly enriched (p < 0.05, q < 0.25) in TAM subtypes from ICB PR patients compared with ICB SD/PD patients.
(F) Violin and boxplots comparing expression distributions of immune checkpoint and evasion genes between all TAM from ICB-exposed versus ICB-naive patients. Significance of differential expression (q value) was determined by two-sided Wilcoxon rank-sum test with Bonferroni FDR correction. Boxplots include centerline, median; box limits, upper and lower quartiles; and whiskers extending at most 1.5× the interquartile range past upper and lower quartiles.
(G) Violin and boxplots comparing expression distributions of immune checkpoint and evasion genes between all TAM from ICB PR versus ICB SD/PD patients. Significance of differential expression (q value) was determined by two-sided Wilcoxon rank-sum test with Bonferroni FDR correction. Boxplots include centerline, median; box limits, upper and lower quartiles; and whiskers extending at most 1.5× the interquartile range past upper and lower quartiles.
∗∗∗q < 0.001, ns: not significant, two-sided Wilcoxon rank-sum test with Bonferroni FDR correction. See also Figure S4, Table S3.