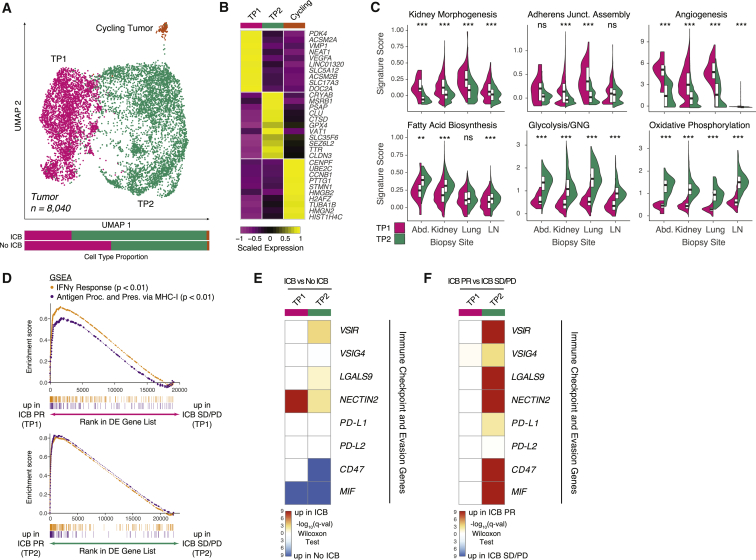
Figure 4.
Two malignant cell programs with distinct metabolic and immune-reactive characteristics are conserved across biopsy sites
(A) UMAP of malignant cells captured across all lesions, colored and labeled by cluster. Bar plots show cluster proportions grouped by ICB treatment history.
(B) Heatmap of scaled normalized expression for cluster-defining genes as determined by two-sided Wilcoxon rank-sum test with Bonferroni FDR correction (q < 0.01).
(C) Violin and boxplots comparing single-cell signature score distributions between the two dominant malignant cell clusters, partitioned by biopsy site. Significance of differential signature enrichment (p value) was determined by two-sided Wilcoxon rank-sum test. Boxplots include centerline, median; box limits, upper and lower quartiles; and whiskers extending at most 1.5× the interquartile range past upper and lower quartiles.
(D) GSEA of hallmark interferon-γ response and gene ontology antigen presentation and processing via MHC class I signatures in TP1 cells from ICB PR patients compared with ICB SD/PD patients (top) and TP2 cells from ICB PR patients compared with ICB SD/PD patients (bottom).
(E) Heatmap of differential expression q values (two-sided Wilcoxon rank-sum test with Bonferroni FDR correction) for immune checkpoint and evasion genes in comparisons of cells within each cluster from ICB-exposed versus ICB-naive patients.
(F) Heatmap of differential expression q values (two-sided Wilcoxon rank-sum test with Bonferroni FDR correction) for immune checkpoint and evasion genes in comparisons of cells within each cluster from ICB PR versus ICB SD/PD patients.
∗∗p < 0.01, ∗∗∗p < 0.001, two-sided Wilcoxon rank-sum test. See also Figure S5, Table S4.