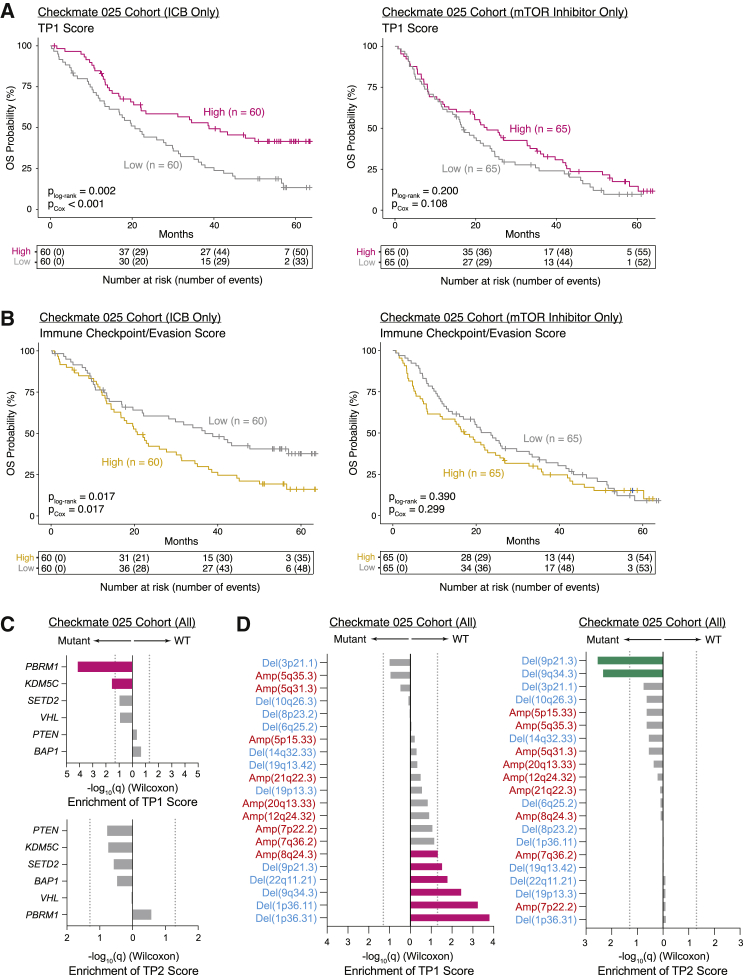
Figure 5.
Tumor program signatures are prognostic in the Checkmate 025 RCC cohort and associated with distinct genomic features
(A) Kaplan-Meier analysis of overall survival (OS) in the Checkmate 025 RCC cohort, with patients separated by high and low TP1 score in bulk RNA-seq. plog-rank, log-rank test p value; pCox, p value determined via a multivariate Cox proportional hazard model using TP1 score dichotomized within treatment arm and incorporating age, sex, MSKCC risk group, prior lines of therapy (≤1 or ≥2), and days between biopsy collection and start of trial therapy as covariates.
(B) Kaplan-Meier analysis of OS in the Checkmate 025 RCC cohort, with patients separated by high and low immune checkpoint/evasion score in bulk RNA-seq. plog-rank, log-rank test p value; pCox, p value determined via a multivariate Cox proportional hazard model using immune checkpoint/evasion score dichotomized within treatment arm and incorporating age, sex, MSKCC risk group, prior lines of therapy (≤1 or ≥2), and days between biopsy collection and start of trial therapy as covariates.
(C) Bar plots comparing TP1 and TP2 score between mutant and wild-type samples for commonly mutated genes in the Checkmate 025 RCC cohort. Significance of differential score enrichment (q value) determined by two-sided Wilcoxon rank-sum test with Benjamini-Hochberg FDR correction. Gray dotted line corresponds to q = 0.05.
(D) Bar plots comparing TP1 and TP2 score between mutant and wild-type samples for common copy number alterations in the Checkmate 025 RCC cohort. Significance of differential score enrichment (q value) determined by two-sided Wilcoxon rank-sum test with Benjamini-Hochberg FDR correction. Gray dotted line corresponds to q = 0.05.