Figure 4.
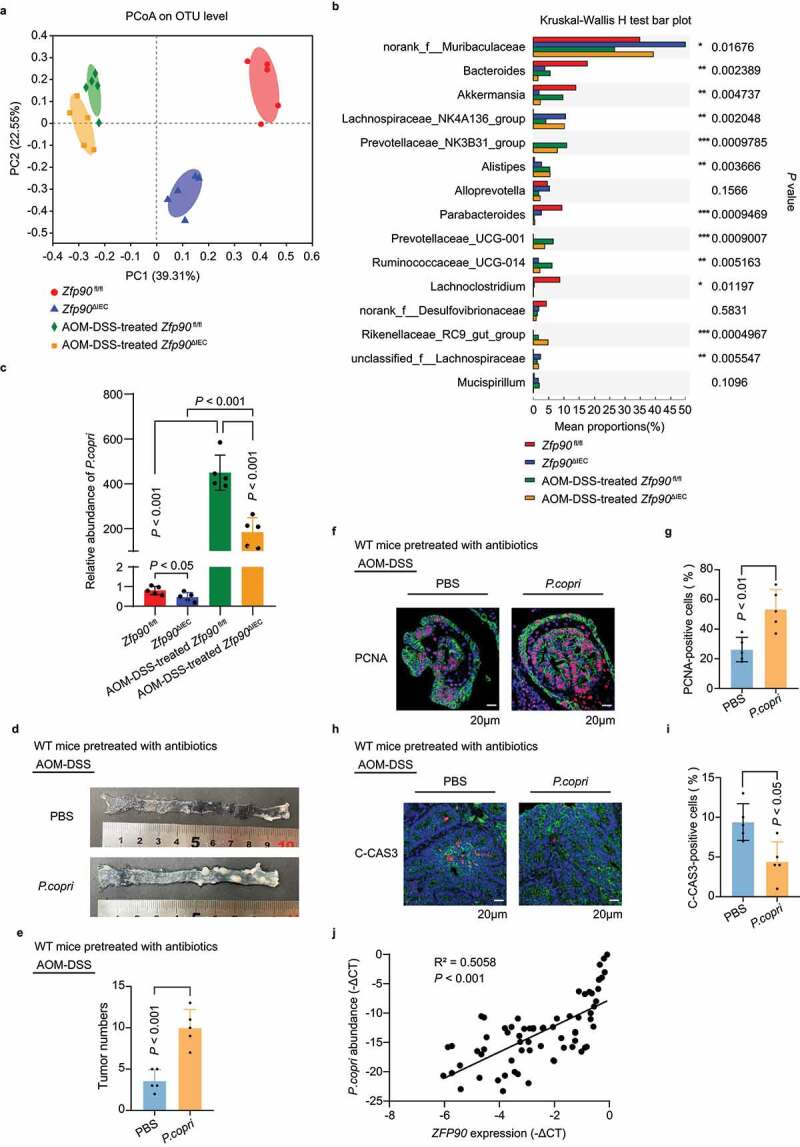
The gut microbiota is altered by intestine-specific deletion of oncogene Zfp90. a Principal component analysis plot based on bacterial 16S ribosomal DNA gene sequences of the fecal content from Zfp90fl/fl and Zfp90ΔIEC mice before and after AOM-DSS-treatment (n = 5 per group). b Differential abundances of bacteria in the fecal content of Zfp90fl/fl and Zfp90ΔIEC mice before and after AOM-DSS-treatment. c Real-time PCR was performed to determine the abundance of P. copri in the fecal content of Zfp90fl/fl and Zfp90ΔIEC mice before and after AOM-DSS-treatment. d Representative images of colons in PBS- and P. copri-treated WT mice after AOM-DSS treatment. e Tumor numbers in PBS- and P. copri-treated WT mice after AOM-DSS treatment. f Representative immunofluorescence staining of the PCNA protein in tumor tissues from PBS- and P. copri-treated WT mice after AOM-DSS treatment. Sections were stained with DAPI (blue), CDH1 (green) and PCNA (red). g The percentage of PCNA-positive tumor cells was quantified. h Representative immunofluorescence staining of C-CAS3 protein in tumor tissues from PBS- and P. copri-treated WT mice after AOM-DSS treatment. Sections were stained with DAPI (blue), CDH1 (green) and C-CAS3 (red). i The percentage of C-CAS3-positive tumor cells was quantified. j The correlation between P. copri abundance and ZFP90 expression in specimens from patients with CRC. Statistical analysis was performed using nonpaired two-tailed t-test (e, g, i), one-way ANOVA (c), and Wilcoxon’s rank-sum test (j)
