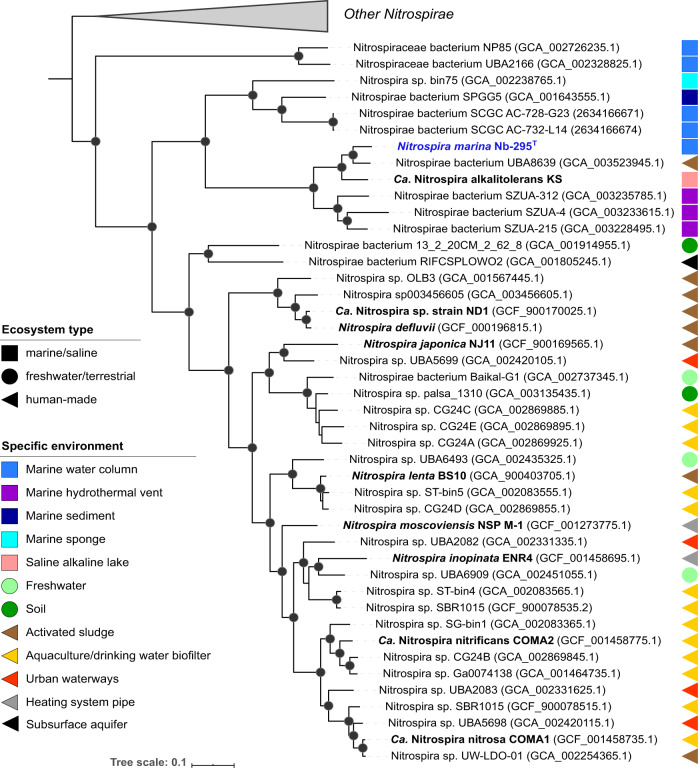
Fig. 1. Maximum likelihood phylogenetic tree of representatives of the phylum Nitrospirae.
The multiple sequence alignment consisting of 120 concatenated phylogenetic marker genes contained 95 genomes and metagenome-assemble genomes (MAGs) from the Genome Taxonomy Database (GTDB) (Release 04-RS89, 19 June 2019), the genome of N. marina, the MAG of Ca. N. alkalitolerans and two open ocean single-cell amplified genomes (SAGs), AC-738-G23 and AC-732-L14. All MAGs and SAGs were estimated to be ≥50% complete with ≤5% contamination. Nodes with UFBoot support of at least 95% are indicated as black filled circles. Cultured representatives are shown in bold. The scale bar represents 0.1 substitutions per site.