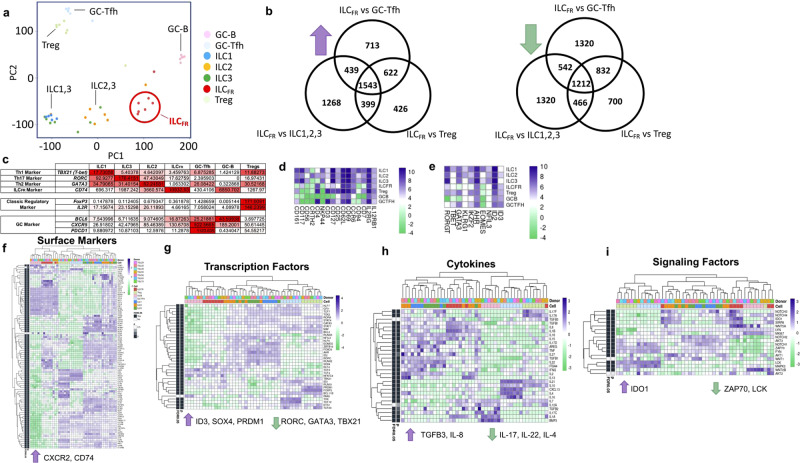
Fig. 2. ILCFR are distinct from other ILC subsets, Tregs, and GC-B or GC-Tfh cells.
RNAseq analysis for GC-Tfh (light blue), GC-B (pink), Treg (light green), ILC1 (blue), ILC2 (orange), ILC3 (green), and ILCFR (red) cells in healthy human tonsils. mRNA was extracted followed by gene expression profiling by using LIMMA in R Bioconductor software. a Principle component analysis of 40,000 unsupervised genes from the indicated cell populations. n = 7 biologically independent tonsils. b Venn diagram analysis of upregulated (left) or downregulated (right) differentially expressed genes comparing ILCFR population to Treg, ILC1, 2 and 3, or GC-Tfh. n = 7 biologically independent tonsils. RPKM values were used to show major transcription factors from ILC subtypes, and important germinal center localization makers in c with a table demonstrating that ILCFR are a distinct cell subtype and are localized to germinal center. d RNAseq analysis of ILCFR for selected common ILC surface markers and e transcription factors. Genes related to innate and adaptive immunity were selected for heatmap analysis for f Surface markers, g Transcription Factors, h Cytokines, and i Signaling Factors. RNA of ILCFR was extracted with RNeasy Micro Kit (Qiagen) and analyzed by NextSeq 550 (Illumina) sequencing on a High Output flow cell using a 75 base pairs, Paired End run. Human TMNCs n = 6 or 7 biologically independent tonsils averaged together for relative expression.