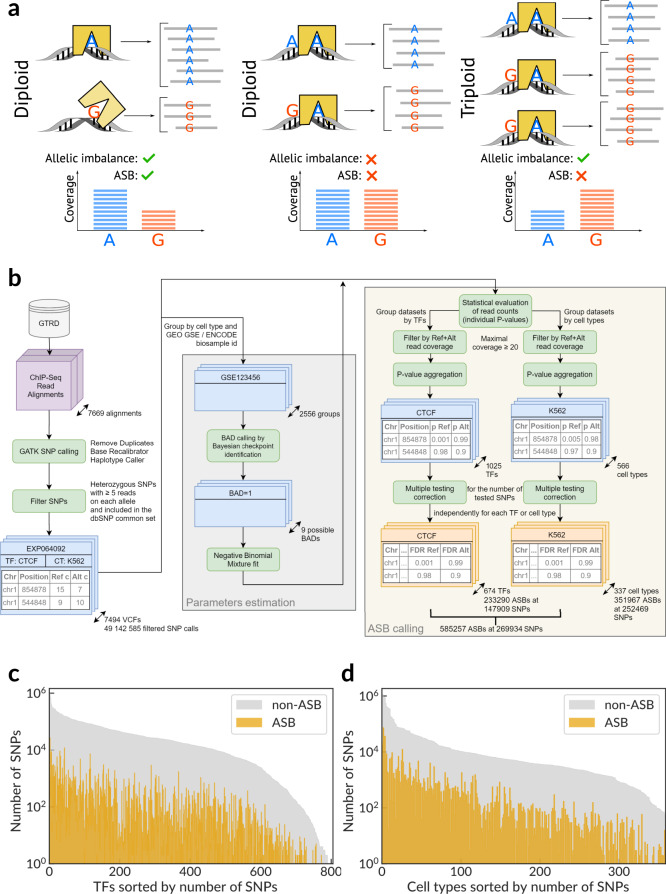
Fig. 1. A scheme of allele-specific binding events, an overview of the ADASTRA pipeline, and its application to ChIP-Seq data.
a ChIP-Seq data allow detecting ASB events by estimating the imbalance of reads carrying alternative alleles. ASBs must be distinguished from sites where the allelic imbalance is caused by aneuploidy and copy-number variants. b The scheme of the ADASTRA pipeline: variant calling in read alignments from GTRD, estimation of statistical model parameters and background allelic dosage, filtering, and statistical evaluation of candidate ASBs. ADASTRA generates two complementary data sets: transcription factor ASBs (pairs of an SNP and a TF) and cell type ASBs (pairs of an SNP and a cell type). SNPs are annotated according to dbSNP IDs. c, d Number of SNPs (dbSNP IDs, Y-axis) with significant ASB events for various transcription factors and for various cell types. TFs or cell types (X-axis) are sorted by the number of SNPs. SNP single-nucleotide polymorphism, BAD background allelic dosage, ASB allele-specific binding, GTRD gene transcription regulation database, ADASTRA Allelic Dosage-corrected Allele-specific human Transcription factor binding sites.