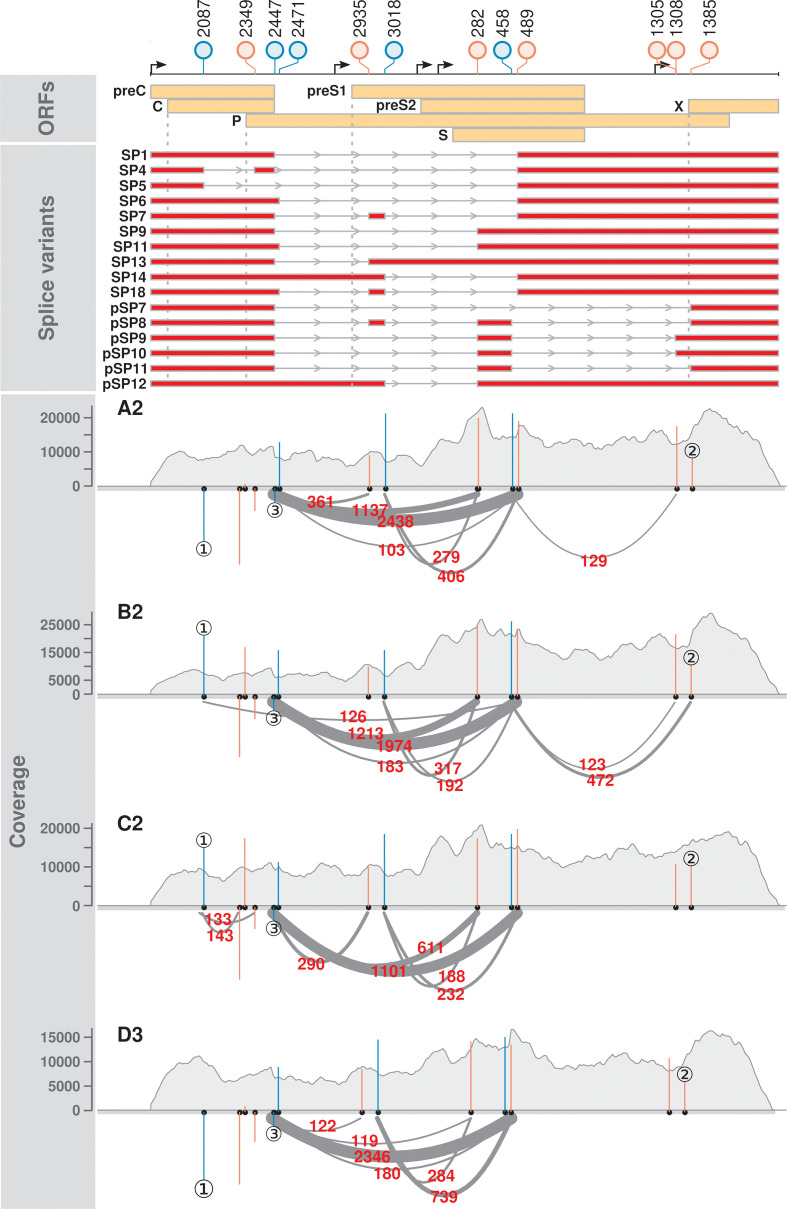
Fig. 3.
Distinct splicing profiles were observed across the HBV genotypes. The lollipop plot indicates the positions of splice sites relative to the EcoRI site of genotype C2. Blue and red colours indicate 5′ and 3′ splice sites, respectively. SP and pSP denote the known and putative spliced pgRNA transcripts, respectively (splice variants panel). These splice variants were reproducibly detected across the independent biological replicates of HBV-transfected Huh7. Grey dotted lines denote the positions of initiation codons of C, P, preS1 and X reading frames (ORFs panel). Read coverage is shown in grey (coverage panel). Arcs represent RNA-seq reads mapped across the splice junctions (supporting read counts in red colour). Only the splice junctions supported by ≥100 reads are shown for readability purposes. Blue and red vertical lines indicate the MaxEntScan scores of the 5′ and 3′ splice sites, respectively (coverage panel). A positive MaxEntScan score predicts a good splice site sequence context, whereas a negative score predicts a poor splice site sequence context. Three main scenarios were observed. ① The presence and absence of spliced reads at position 2087 were predicted by MaxEntScan scores, in which reads were found to map across the 5′ splice sites with strong positive scores (B2 and C2), but not those with strong negative scores (A2 and D3). ② Varying spliced read counts could not be explained by similar scores. ③ Most spliced reads were mapped across a weak splice donor site. See also Fig. S4, Table S8.