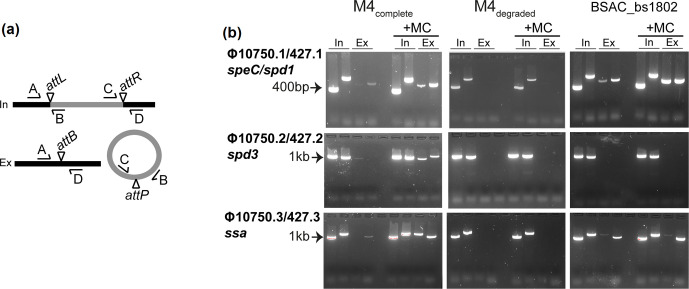
Fig. 3.
Phage induction in M4complete isolates but not M4degraded isolates. (a) Primers were designed to detect integrated (In) and excised (Ex) prophage. Primer pairs A+B and C+D spanned the attachment sites attL and attR, respectively, formed when the prophage (grey) is integrated into the chromosome (black). Primer pairs A+D and C+B spanned the attachment site attB on the bacterial chromosome and attP on the prophage, respectively, detecting prophage excision. (b) Both integrated prophage, as indicated by bands in the two ‘In’ lanes, and excised prophage, as indicated by bands in the two ‘Ex’ lanes (primer pairs A+D and C+B) were detected in M4complete for all three prophages. Excision was enhanced by the addition of mitomycin C (+MC). No excision was detected for all three prophages in M4degraded, as indicated by bands only in the In lanes but not the Ex lanes, even with mitomycin C. The exception was BSAC_bs1802, where excision of Φ10750.1/427.1 and Φ10750.3/427.3 was detected but excision of Φ10750.2/427.2 was not. Representative gels are shown for single isolates out of the five M4complete BSAC isolates tested (BSAC_bs1388) and four M4degraded tested (BSAC_bs472). Primer pairs were used in the following order: In lane 1, A+B (attL); In lane 2, C+D (attR); Ex lane 3, A+D (attB); Ex lane 4, C+B (attP).