Fig. 4. Myo15 positions Mical to spatially target and expand F-actin disassembly.
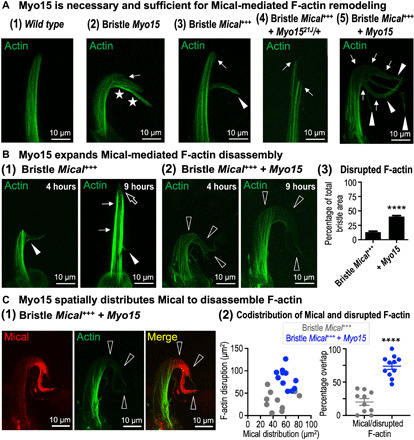
Genotypes are as previously described plus the addition of GFPactin transgene to visualize actin. (A) Myo15 is necessary and sufficient for Mical-mediated F-actin disassembly. (1) Normal bristle F-actin organization (B11-GAL4/+, UAS:GFPActin/+). (2) Elevating Myo15 induces regions of more (stars) and less (arrow) F-actin. Abnormal bristle bends, and other alterations accompanied these F-actin changes (e.g., stars; see also figs. S2, E and F, and S6A). (3) Elevating Mical induces regions of less F-actin (arrow) and branching (arrowhead) (4, 7). (4) Reducing Myo15 decreases Mical-triggered F-actin disassembly and branching (arrows). (5) Elevating Myo15 increases Mical-triggered F-actin disassembly (arrows) and branching (arrowheads). (B) Myo15 expands Mical-mediated F-actin disassembly. Bristle development tracked as in Figs. 2D [(1) and (2)] and 3C (2) and fig. S9B. (1) Elevating Mical induces F-actin disruptions and branching (4 hours, closed arrowhead). Bristles then extend past this branch point (9 hours, thin arrows), where F-actin disruptions (open arrow) and branching [13 hours; Fig. 2D, (1)] occur again. (2) Elevating Myo15 increases Mical-triggered F-actin disruptions (4 hours, open arrowheads), and these further increase with age (9 hours, open arrowheads). (3) Percentage of the bristle with disrupted F-actin. Means ± SEM. n ≥ 12 bristles (6 to 12 animals) per genotype. ****P < 0.0001, unpaired t test (two-tailed). (C) Myo15’s increased broadening of Mical’s distribution (red) overlaps with Myo15’s increased broadening of Mical-triggered F-actin (green) disruptions [open arrowheads in (1) and quantified in (2)]. Means ±SEM. n > 10 bristles (5 to 10 animals) per genotype. ****P < 0.0001, two-tailed t test.
