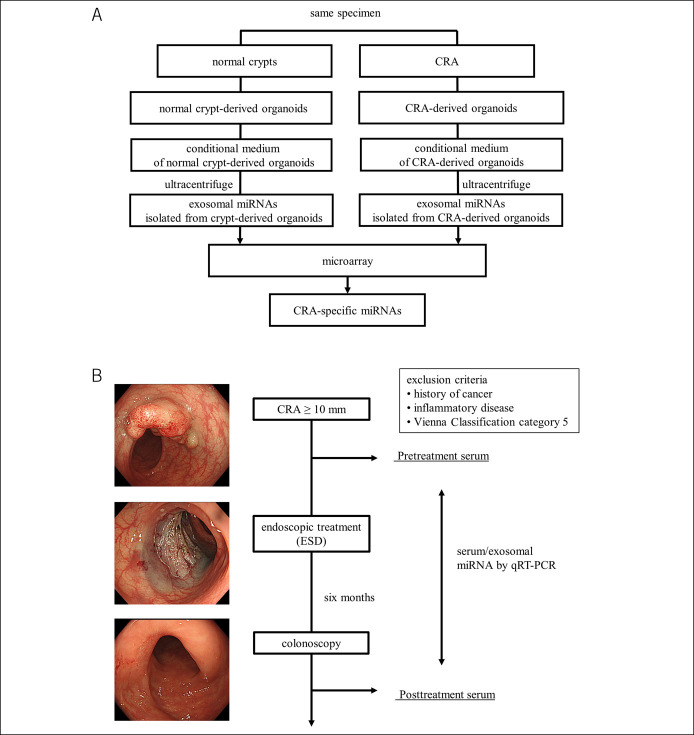
Figure 1.
Study design overview. (a) Selection of candidate miRNAs using the organoid culture. Organoids from the normal colon crypts and CRA are established in pairs from endoscopically resected specimens. Then, the exosomes are isolated from conditional medium collected from each organoid using ultracentrifugation. Then, microarray analysis is conducted for each of the exosomal miRNAs isolated from the normal colon crypt-derived organoids and CRA-derived organoids. CRA-specific miRNA candidates are extracted. (b) Flow diagram showing the strategy for qRT-PCR analysis of serum miRNAs before and after endoscopic resection. Pretreatment sera are obtained from patients with CRA. Then, CRA is resected using ESD within 1 month. Posttreatment sera are obtained 6 months after resection. At that time, total colonoscopy has been performed to confirm healing of the mucosa and lack of recurrence or residual tumor in the treated area. The expression changes between pretreatment sera and posttreatment sera of candidate miRNAs are validated using RT-PCR. CRA, colorectal adenoma; miRNAs, microRNAs; ESD, endoscopic submucosal dissection; qRT-PCR, quantitative reverse transcription-polymerase chain reaction.