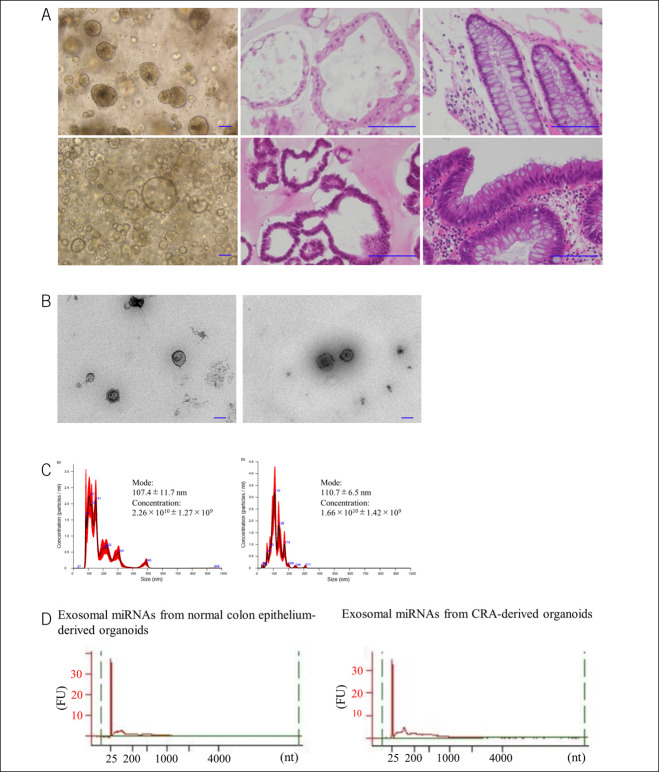
Figure 2.
Patient-derived organoids and isolation of exosomes. (a) Top panels show the images of normal colon crypts. Optical microscope image of organoids (left), H&E staining of organoids (center), and H&E staining of endoscopically resected specimens (right). Bottom panels show the images of the CRA. Optical microscope image of organoids (left), H&E staining of organoids (center), and H&E staining of endoscopically resected specimens (right). Each scale bar is 100 μm. (b) Electron microscope imaging of the isolated exosomes. Images of exosomes from normal colon crypt-derived organoids (left) and of exosomes from CRA-derived organoids (right). Original magnification: ×15,000. Scale bar shows 100 nm. (c) Size distribution of isolated exosomes analyzed using a Nanosight NS300. Size distribution of normal colon crypt-derived organoids (left). Size distribution of exosome from CRA-derived organoids (right). (d) Exosomal miRNAs are prepared from normal crypt-derived and CRA-derived organoids. Their integrity and quality are examined using a bioanalyzer system. Exosomal miRNA did not include ribosomal RNAs. Image of exosomal miRNAs from normal colon crypt-derived organoids (left), image of exosomal miRNAs from CRA-derived organoids (right). CRA, colorectal adenoma; H&E, hematoxylin–eosin; miRNA, microRNA.