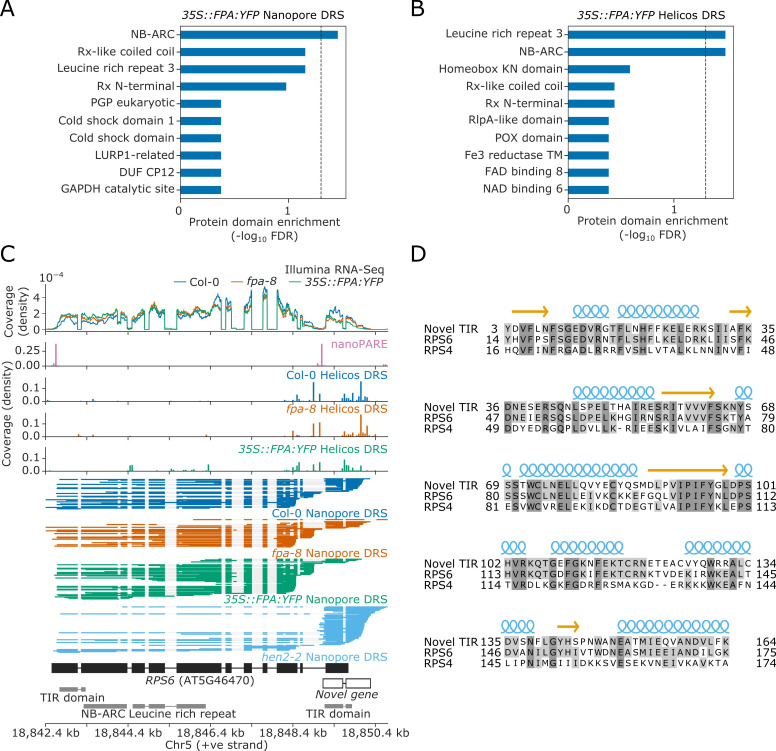
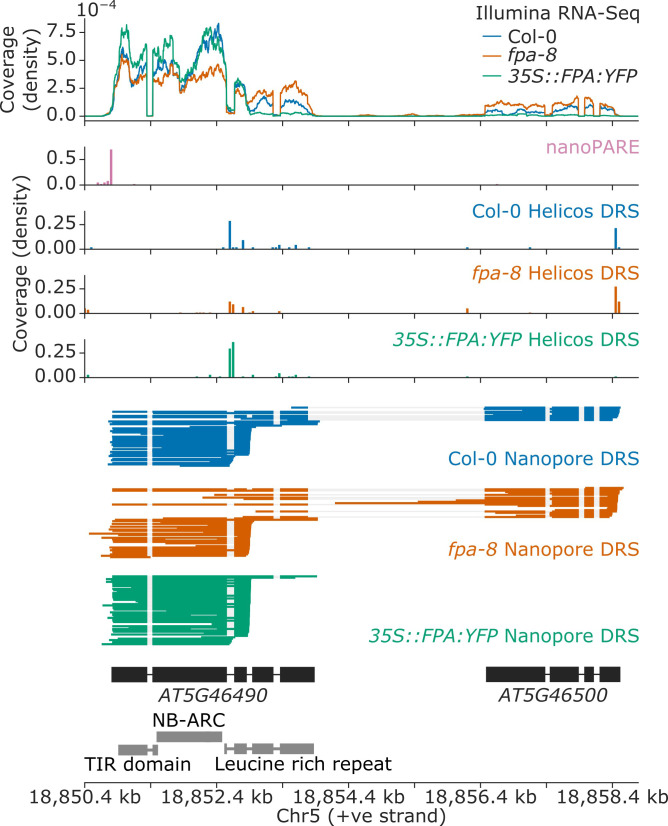
Figure 3. Nanopore and Helicos DRS identify NLR genes regulated by alternative polyadenylation.
(A–B) Protein domain enrichment analysis for loci with increased proximal poly(A) site selection in 35S::FPA:YFP line, as detected using (A) Nanopore DRS or (B) Helicos DRS. (C) Nanopore DRS reveals the complexity of RNA processing at RPS6. Protein domain locations (shown in grey) represent collapsed InterPro annotations. The novel TIR domain was annotated using InterProScan (Mitchell et al., 2019). (D) Protein alignment of the predicted TIR domain from the novel gene downstream of RPS6, with the sequence of the TIR domains from RPS6 and RPS4. Helix and strand secondary structures (from UniProt: RPS4, Q9XGM3) are shown in blue and yellow, respectively. Residues are shaded according to the degree of conservation.