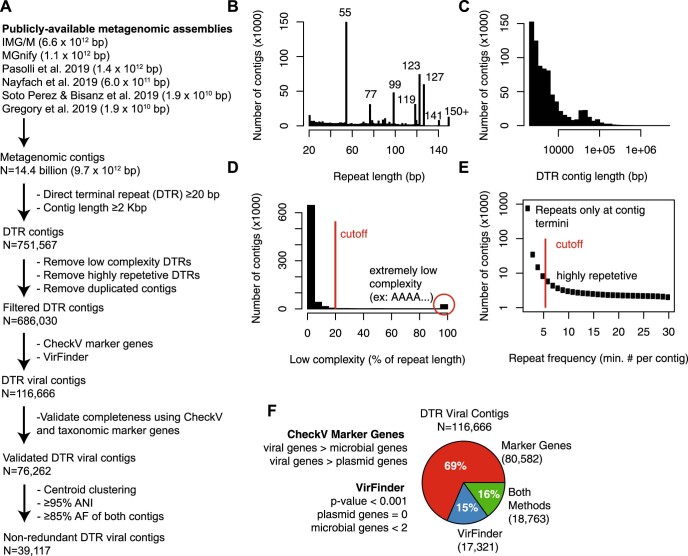
Extended Data Fig. 3. Identification of DTR viral contigs.
a) Publicly available metagenomes were systematically mined for 76,262 DTR viral contigs, resulting in 39,117 non-redundant contigs after de-replication at 95% ANI over 85% the length of both sequences. b–e) Summary statistics across the 751,567 DTR contigs before filtering. b) Distribution of the length of direct terminal repeats (DTRs). A considerable number of DTRs occur at specific lengths (for example 55, 77, 99 bp). These odd-numbered lengths likely correspond with k-mer lengths utilized by various metagenomic assembly tools. When faced with assembling reads from a circular template, they appear to break the contig in a random location and leave behind a repeated sequence at the start and end of the contig equal to the k-mer length. c) The length (log scale) of all DTR contigs. d) A small number of contigs are likely false positives due to a low complexity repeat (for example AAAAAA…) or e) a highly repetitive repeat (that is occurring not just at termini). f) After removing potentially spurious complete genomes, the DTR contigs were screened for viral signatures, revealing 116,666 viral contigs. These were identified using a combination of CheckV’s marker genes, plasmid genes from recent publications, and VirFinder.