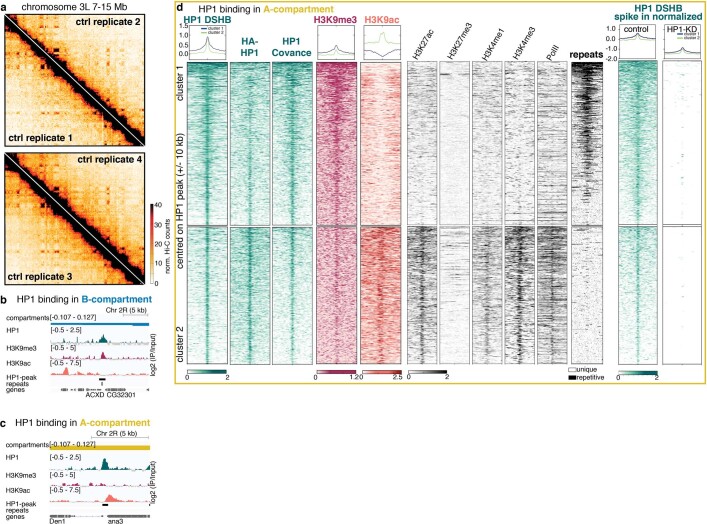
Extended Data Fig. 2. Characterization of HP1 binding within A- and B-compartment.
a, Hi-C contact maps with contact frequencies of chromosome 3L (7–15 Mb) at a resolution of 40 kb. Four out of seven biological replicates are shown. b, Representative example of HP1 binding in a B-compartment region. c, Representative example of HP1 binding in an A-compartment region. d, Extended characterization of HP1 binding in A-compartment regions. The heat maps show ChIP–seq signal and repeat coordinates in ±10 kb centred around HP1 peaks. Figure 2d shows only cluster 2 containing HP1 peaks that localize within non-repetitive, active regulatory sequences enriched in H3K9ac, H3K27ac, H3K4me1/3 as well as polymerase II. We validated this cluster in active regions by performing ChIP–seq with different antibodies against HP1 (HA antibody against a Flag-HA-HP1-tagged transgene (second heat map) and HP1 Covance antibody (third heat map)). We further performed ChIP–seq in HP1-KD embryos using λ-DNA spike-in to normalize the signal and found a strong reduction of HP1 binding. This further validates the specificity of the HP1 peaks. See Supplementary Methods for further information. A second cluster of HP1 binding events (cluster 1) occurs in repetitive chromatin regions that are largely devoid of active histone modification marks.