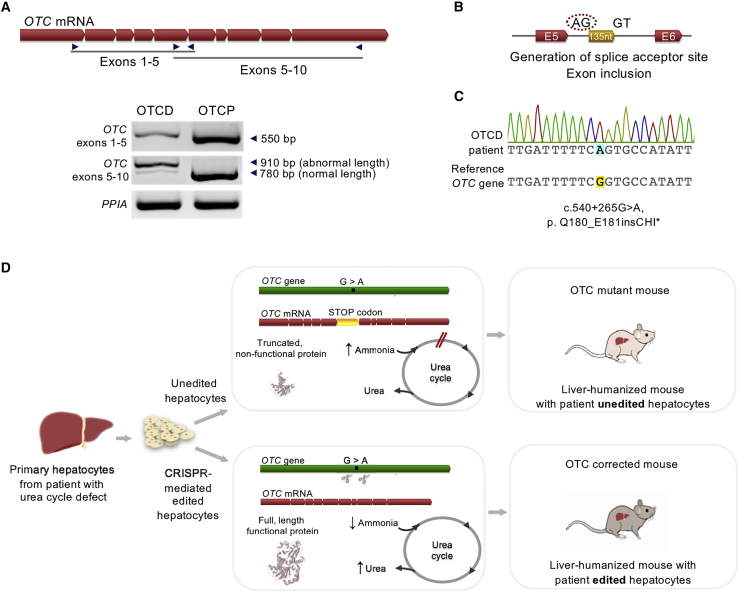
Figure 1.
Disease-causing variant identification and study outline
(A) Amplification of OTC transcript in hepatocytes from OTC-deficient (OTCD) patient and OTC-proficient (OTCP) donor. Red arrow-shaped boxes represent exons of the OTC transcript, which were amplified with PCR primers indicated, spanning exons 1–5 and 5–10. The length of each amplicon product is indicated with arrows. Peptidylprolyl isomerase A (PPIA) transcript was used as an endogenous control. (B) Sequencing of patient OTC transcript revealed a 135-nt sequence inclusion, identical to an intronic region, between exons 5 and 6. Genomic structure of exons 5 and 6 of the OTC gene and the intronic region included in the transcript are shown. Red arrow-shaped boxes and horizontal lines represent exons and introns, respectively. Dinucleotides of the generated splice acceptor (AG) and the naturally existing donor (GT) flank the 135-nt insertion (yellow box). (C) Sequencing of OTC intronic region containing the mutation and alignment to reference gene (NCBI Gene: 5009). The genetic mutation (c.540+265 G > A) results in the generation of a splice acceptor (AG) site inserting an intronic region in the OTC transcript and generating a premature stop codon shortly after exon 5 (p. Q180_E181insCHI∗). (D) Graphical depiction of the mutation identified, the consequences of the mutation, and the corrective strategy, followed by the characterization of the phenotype of the liver-humanized mice generated with unedited and edited cells.