Figure 2.
Genetic correction and in vitro phenotype characterization
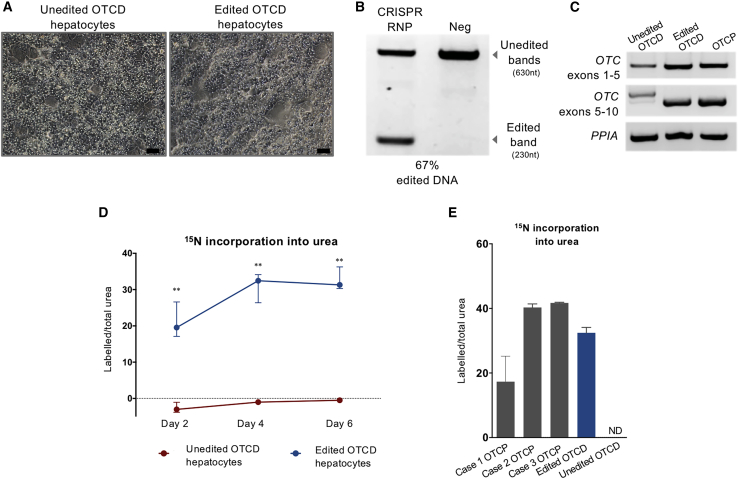
(A) Phase contrast microscopy imaging of unedited or edited OTCD hepatocytes in vitro 2 days post-electroporation. Scale bar, 50 μm. (B) PCR amplification of targeting region in edited and unedited OTCD hepatocytes. Upper and lower bands indicate unedited (wild type) and edited DNA, respectively. Cleavage efficiency was estimated to be 67% based on band intensity and amplicon length. (C) PCR amplification of OTC transcript in unedited or edited OTCD hepatocytes, as well as OTCP cells. Transcripts of 2 lengths were observed in unedited OTCD hepatocytes, with a >100-nt difference, which corresponds to the intronic region included in the transcript. PPIA transcript was used as an endogenous control. (D) Phenotypic characterization in vitro assessed through 15N incorporation into urea. Unedited and edited OTCD hepatocytes were incubated with 15NH4Cl. Ratios of labeled to total urea produced by the cells on days 2, 4, and 6 post-electroporation are presented in the graph. Values and errors represent medians and interquartile ranges (unedited cells n = 6, edited cells n = 6 on each day). Statistical analysis was performed using the Mann-Whitney U test. Day 2, p = 0.0043; day 4, p = 0.0022; and day 6, p = 0.0022. (E) Phenotypic characterization in vitro assessed through 15N incorporation into urea. Ratios of labeled to total urea produced by edited and unedited OTCD hepatocytes, as well as primary hepatocytes from 3 OTCP donors assessed on day 4 of culture, are shown. Values and errors represent medians and interquartile ranges (OTCP cases n = 3; edited and unedited OTCD cells n = 6). ND, not detectable.