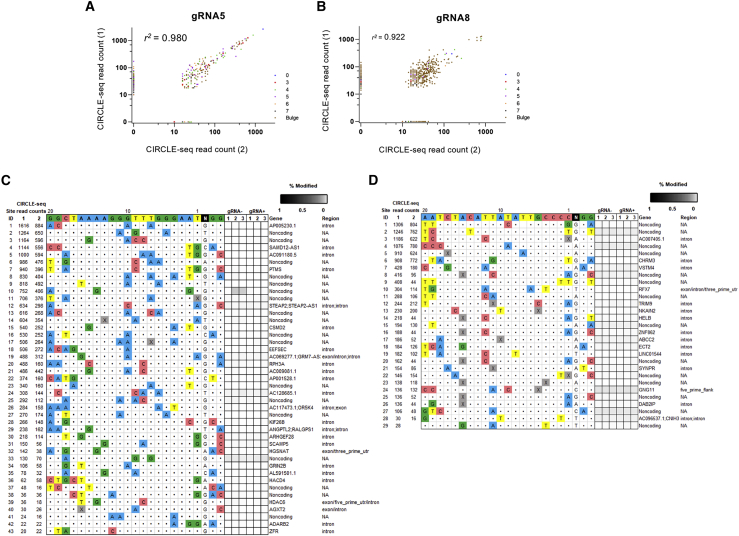
Figure 6.
Off-target editing
(A and B) Off-target discovery of OTC-targeting gRNAs by CIRCLE-seq. Scatterplots of CIRCLE-seq read counts for off-target cleavage sites identified in vitro with Cas9-gRNA5 (A) or Cas9-gRNA8 (B) in 2 replicates on genomic DNA from patient-derived primary hepatocytes. Read counts are shown on a logarithmic scale. Each site is color coded for the number of mismatches relative to the on-target site. Correlation r2 values in between 2 replicates were obtained by Pearson correlation performed using all values shown in the scatterplot. (C and D) Assessment of off-target indels induced by Cas9-gRNA5 (C) or Cas9-gRNA8 (D) in patient-derived primary hepatocytes. Indel frequencies as determined by targeted amplicon sequencing are presented as heatmaps for the off-target sites (identified from CIRCLE-seq experiments). CIRCLE-seq read-count numbers for each site are listed. Mismatches relative to the on-target site are shown with colored boxes, whereas bulges are indicated with X. Each locus was assayed in n = 3 transfection replicates (labeled 1, 2, and 3) using genomic DNA isolated from the cells electroporated with RNP complex Cas9-gRNA (gRNA+) or control-only Cas9 protein (gRNA−). Genomic identifications and locations for each locus are presented. NA, not applicable.