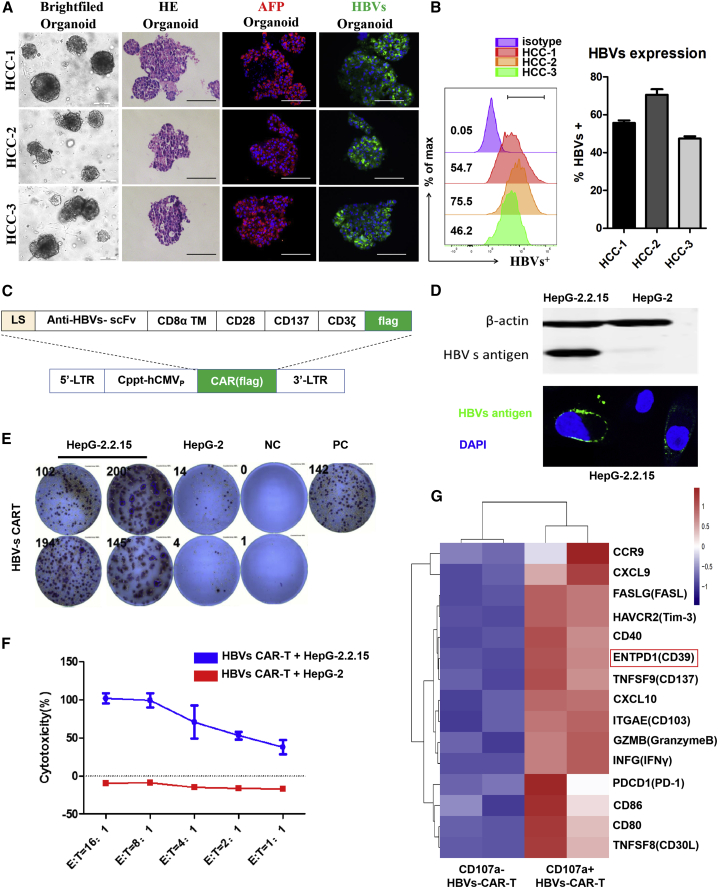
Figure 1.
HBVs protein was abundant on HCC organoids, and HBVs-CAR-T cells were constructed for cellular immunotherapy
(A) Bright-field microscopy images, H&E staining, and immunofluorescence staining of HCC markers AFP (red) and HBVs (green) for organoids were shown separately. Nuclei were stained with DAPI (blue). Scale bars, 200 μm (bright field), 100 μm (H&E and immunofluorescence). (B) Flow cytometry was used to analyze the frequency of HBVs protein expression on HCC organoids of 3 patients. (C) Schematic representation of the lentiviral vectors carrying a HBVs-specific 3rd-generation CAR moiety. (D) Western blotting and immunofluorescence staining for HBVs in HepG-2.2.15 cells; HepG-2 cells served as the mock cells. (E) Immune response was evaluated by measuring the number of IFN-γ production and spot-forming cells (SFCs) per 104 HBVs-CAR-T cells (E:T = 4:1). CAR-T cells added with PHA served as PC (positive control) and CAR-T cells alone as NC (negative control). (F) Cytotoxic activity of HBVs-CAR-T cells on HepG-2.2.15 targets cells were detected by LDH assay (n = 3, 3 healthy donors). (G) After co-culture with HepG-2.2.15 cells for 24 h (E:T = 4:1), CD107a+ and CD107a− HBVs-CAR-T cells were obtained by flow sorting, and their transactional profiling was analyzed by RNA sequencing (RNA-seq). Heatmap of RNA-seq showed the expression of genes involving in cytotoxicity, adhesion, and chemotaxis in different effector sub-populations, with p < 0.01. RNA expression levels were indicated with a red/blue scale for high and low expression levels, respectively.