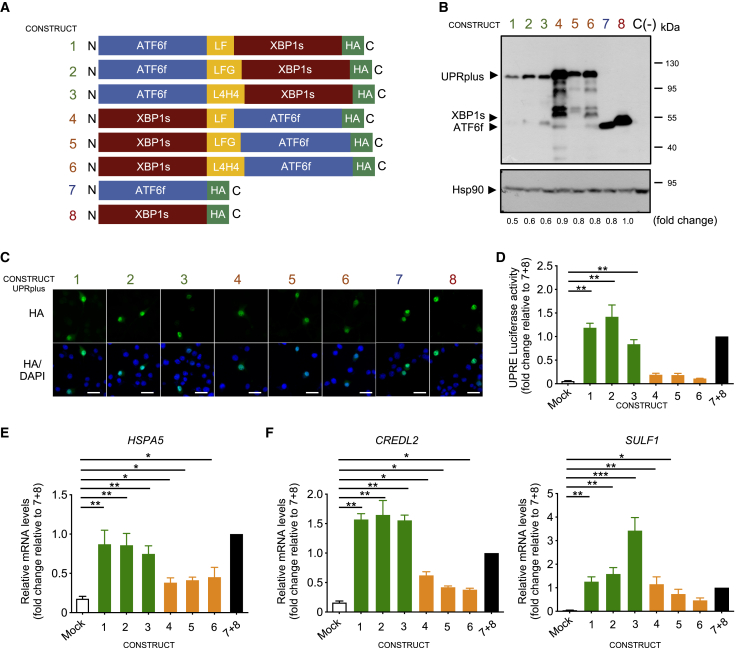
Figure 1.
Generation of ATF6f and XBP1s fusion constructs
(A) Diagram of AAV constructs generated to deliver active UPR transcription factors. Different artificial heterodimers were generated by fusing ATF6f and XBP1s using three different linker sequences (yellow boxes) by combining their positions in the C-terminal and N-terminal regions. All constructs contain an HA tag at the C-terminal region (green box) for detection of the expression of the transgene. (B) HEK293T cells were transiently transfected with the six fusion constructs described in (A), in addition to XBP1s-HA or ATF6f-HA alone and empty vector C (–). After 48 h of expression, cell extracts were analyzed by western blot using an anti-HA antibody. Hsp90 was monitored as a loading control. Fold changes are shown related to XBP1s expression levels. (C) In parallel, cells described in (B) were analyzed by immunofluorescence using an anti-HA antibody (green). Co-staining with the nuclear marker Hoechst (blue) was performed. Scale bars, 10 μm. (D) HEK293T cells were transiently co-transfected with the six variants of pAAV-UPRplus or single constructs together with the UPRE-luciferase reporter and Renilla constructs. After 48 h luciferase activity was measured using a luminometer. (E and F) HEK293 cells were transiently transfected with the six versions of UPRplus or control vectors. After 48 h, the mRNA levels of the indicated UPR-target gene were measured by real-time RT-PCR. All samples were normalized to β-actin levels. mRNA levels are expressed as fold increase over the value obtained in control cells transfected with an equivalent 1:1 mixture of individual XBP1s and ATF6f expression vectors. In (D) and (E), the mean and standard error are presented for three independent experiments. Statistical analysis was performed using Dunnett’s multiple comparison test. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.