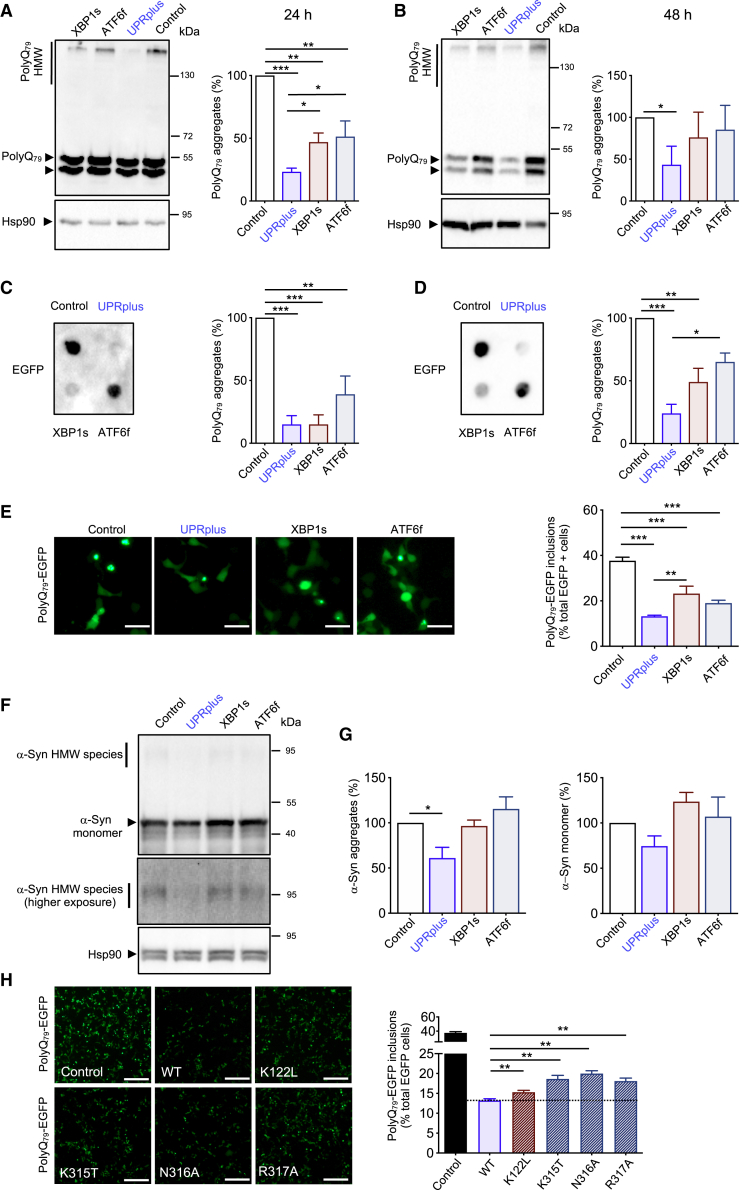
Figure 3.
UPRplus expression reduces mutant huntingtin and α-synuclein aggregation
(A and B) Neuro2A cells were transiently co-transfected with expression vectors for polyQ79-EGFP and XBP1s, ATF6f, UPRplus, or empty vector (control). After 24 (A) or 48 h (B), polyQ79-EGFP detergent-insoluble aggregates were measured in cell extracts prepared in Triton X-100 by western blot. Levels of Hsp90 were measured as the loading control. Left panel: high molecular weight (HMW) polyQ79-EGFP aggregates were quantified. (C and D) PolyQ79-EGFP detergent-insoluble aggregates were measured by filter trap assay after 24 (C) or 48 h (D) of transfection (right panel). Left panels: polyQ79-EGFP aggregates were quantified. (E) In cells described in (A), polyQ79-EGFP intracellular inclusions were quantified after 48 h of expression by fluorescence microscopy (right panel). Scale bars, 20 μm. Left panel: the number of cells displaying intracellular inclusions was quantified in a total of at least 300 cells per experiment. (F) HEK293T cells were transiently co-transfected with expression vectors for α-synuclein-RFP (α-syn) together with expression vectors for UPRplus, ATF6f, XBP1s, or empty vector (control). After 48 h, α-syn aggregates were measured in cell extracts prepared in 1% Triton X-100 by western blot (upper panel). Middle panel: higher exposure of the upper panel highlighting HMW species of α-syn. Levels of Hsp90 were monitored as the loading control (lower panel). (G) α-Syn-RFP HMW species (left panel) and α-syn monomers (right panel) were quantified. (H) Neuro2A cells were transiently co-transfected with expression vectors for polyQ79-EGFP together with empty vector (control), UPRplus WT, or the UPRplus mutants K122L, K315T, N316A, and R317A. After 48 h, the accumulation of polyQ79-EGFP intracellular inclusions was visualized by fluorescence microscopy (right panel). Scale bars, 100 μm. Left panel: the number of cells displaying intracellular inclusions was quantified in a total of at least 300 cells per experiment. In (A)–(G), the mean and standard error are presented for three independent experiments. In (G), four independent experiments were quantified. Statistical analysis was performed using Dunnett’s multiple comparison test. ∗p < 0.05, ∗∗p < 0.01).