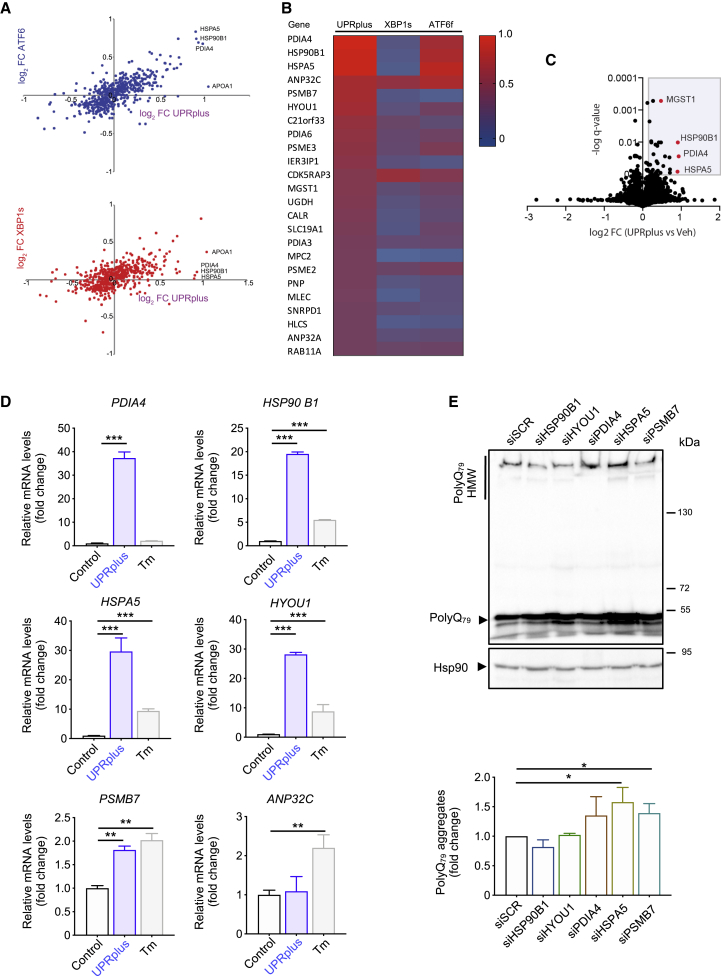
Figure 4.
UPRplus expression remodels proteostasis pathways
(A) Quantitative proteomics was performed in protein extracts from HEK293T cells infected for 48 h with the following viral particles: AAV-UPRplus, AAV-XBP1s, AAV-ATF6f, or AAV-empty (vehicle). The data were analyzed and plotted of log2 fold change (FC) for proteins identified in MuDPIT analysis. Plot showing the correlation between gene expression in cells expressing UPRplus or ATF6f (upper panel) and UPRplus or XBP1s (bottom panel). Only genes whose expression is significantly affected (false discovery rate [FDR] < 0.01) are shown. (B) Heatmap analysis showing differential protein expression patterns in ATF6f, XBP1s, or UPRplus overexpression conditions. Color from red to blue indicates high to low expression. (C) Volcano plot showing the correlation between protein expression of HEK293T cells infected with AAV-UPRplus versus AAV-empty (vehicle) viral particles. Only genes whose expression is significantly affected (FDR < 0.01) are shown. (D) The mRNA levels of selected UPR-upregulated genes were monitored in HEK293T cells infected with AAV-empty (control) or AAV-UPRplus viral particles. After 48 h the relative mRNA levels of the indicated genes were measured by real-time RT-PCR. As a positive control, cells were treated with 1 μg/mL tunicamycin (Tm) for 8 h. All samples were normalized to β-actin levels. mRNA levels are expressed as fold increase over the value obtained in the control condition. (E) HEK293T cells were transiently transfected with siRNA against the six top gene genes upregulated by UPRplus. A scrambled siRNA (siScr) was used as a control. 24 h later cells were transfected with a polyQ79-EGFP expression vector followed by western blot analysis after 24 h of expression. Levels of Hsp90 were analyzed as a loading control. Bottom panel: polyQ79-EGFP high molecular weight (HMW) species were quantified. In (D) and (E), the mean and standard error are presented of three independent experiments. Statistical analysis was performed using Dunnett’s multiple comparison test. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.