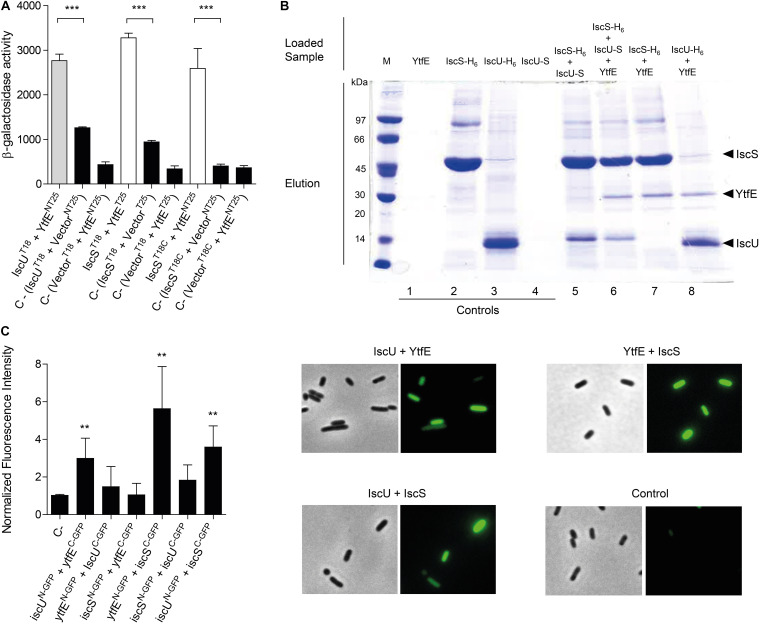
FIGURE 1.
Analysis of the interaction of YtfE with IscS and IscU. (A) The efficiency of the functional complementation between hybrid proteins was measured by β-galactosidase assays. The interaction of YtfE with IscU (gray bar) and IscS (white bars) was evaluated in E. coli cells co-transformed with plasmids containing either iscU or iscS genes cloned at the N- or C-terminal of the T18 fragment (T18/T18C), and the plasmid harboring the ytfE gene cloned at the N- or C-terminal of the T25 fragment (NT25/T25). T25/NT25 empty plasmids together with the T18/18C plasmid containing either the iscU or the iscS gene or T18/18C empty plasmid with NT25/T5 harboring ytfE were utilized as negative controls (black bars). Values are means ± standard errors from at least three independent cultures analyzed in duplicate. ***P < 0.0001; (B) For pulldown assays of YtfE, IscS and IscU, samples were obtained from cell extracts expressing one, two or three proteins previously treated with 1% formaldehyde and loaded in Ni-chelating columns, eluted with 1 M imidazole buffer, and subjected to SDS-PAGE. Lanes 1–4: control samples from extracts expressing only one protein. Lanes 5–8: co-purification of IscS with IscU (5); co-purification of YtfE and IscU with IscS (6); co-purification of IscS with YtfE (7); and co-purification of IscU with YtfE (8); and (C) In BiCF of YtfE and ISC proteins, cells were co-transformed with vectors expressing YtfE and IscS or IscU – GFP fusions. Cells harboring empty plasmids were used as controls. On the left, fluorescence quantification was performed using MetaMorph Microscopy Automation and Image Analysis Software. Fluorescence values for the negative control (empty plasmid vectors) were normalized to 1. **P < 0.005. On the right microscope images (Bright field phase and FITC) of Negative control; iscUN–GFP + YtfEC–GFP; ytfE N–GFP + IscSC–GFP; and iscU N–GFP + IscSC–GFP. Values are means ± standard errors from at least three independent cultures analyzed in duplicate.