Soybean (Glycine max L.) is a valuable food crop that also contributes significantly to soil nitrogen by developing a symbiotic association with nitrogen-fixing rhizobia. Bacterial endophytes (both rhizobial and nonrhizobial) are considered critical for the growth and resilience of the legume host.
KEYWORDS: nonrhizobial endophytes, rhizobial endophytes, soybean rhizosphere microbiome, soybean root nodule microbiome, small-scale spatial soil heterogeneity
ABSTRACT
Soybean root nodules are known to contain a high diversity of both rhizobial endophytes and nonrhizobial endophytes (NREs). Nevertheless, the variation of these bacteria among different root nodules within single plants has not been reported. So far, it is unclear whether the selection of NREs among different root nodules within single plants is a random process or is strictly controlled by the host plant to favor a few specific NREs based on their beneficial influence on plant growth. As well, it is also unknown if the relative frequency of NREs within different root nodules is consistent or if it varies based on the location or size of a root nodule. We assessed the microbiomes of 193 individual soybean root nodules from nine plants using high-throughput DNA sequencing. Bradyrhizobium japonicum strains occurred in high abundance in all root nodules despite the presence of other soybean-compatible rhizobia, such as Ensifer, Mesorhizobium, and other species of Bradyrhizobium in soil. Nitrobacter and Tardiphaga were the two nonrhizobial genera that were uniformly detected within almost all root nodules, though they were in low abundance. DNA sequences related to other NREs that have frequently been reported, such as Bacillus, Pseudomonas, Flavobacterium, and Variovorax species, were detected in a few nodules. Unlike for Bradyrhizobium, the low abundance and inconsistent occurrence of previously reported NREs among different root nodules within single plants suggest that these microbes are not preferentially selected as endophytes by host plants and most likely play a limited part in plant growth as endophytes.
IMPORTANCE Soybean (Glycine max L.) is a valuable food crop that also contributes significantly to soil nitrogen by developing a symbiotic association with nitrogen-fixing rhizobia. Bacterial endophytes (both rhizobial and nonrhizobial) are considered critical for the growth and resilience of the legume host. In the past, several studies have suggested that the selection of bacterial endophytes within root nodules can be influenced by factors such as soil pH, nutrient availability, host plant genotype, and bacterial diversity in soil. However, the influence of size or location of root nodules on the selection of bacterial endophytes within soybean roots is unknown. It is also unclear whether the selection of nonrhizobial endophytes within different root nodules of a single plant is a random process or is strictly regulated by the host. This information can be useful in identifying potential bacterial species for developing bioinoculants that can enhance plant growth and soil nitrogen.
INTRODUCTION
Soybean (Glycine max) is a valuable food crop that also contributes significantly toward soil nitrogen by developing symbiotic associations with nitrogen-fixing rhizobia. Under different soil conditions, soybean can be nodulated by different rhizobial genera, including Bradyrhizobium, Rhizobium, Ensifer (Sinorhizobium), and Mesorhizobium (1–11). The host plant selection of specific rhizobial endophytes is relatively well studied (12–16) and has been reported to be influenced by soil factors, such as pH (6, 17, 18), salinity (3, 13), nutrient availability (19–22), water (16), geographic locations (4), and diversity of the rhizobial community within soil (23, 24). Soil is a highly heterogeneous environment, and its biochemical conditions can greatly vary over short distances (i.e., <1 cm) (25, 26). It is unclear if the same rhizobial species are selected in all root nodules of a plant or if selection of rhizobia in individual root nodules varies depending upon the size or location of the nodules on a root system. Hence one of the objectives of the current study was to assess the potential influence of location or size of a nodule on the selection of rhizobial endophytes within different root nodules of a single plant.
Several studies have observed the presence of various nonrhizobial endophytes (NREs, i.e., bacterial endophytes other than four rhizobial genera) within soybean root nodules (16, 27–41) (see Table S1 in the supplemental material for details). Many culture-based studies have successfully isolated and identified several NREs from root nodules and reported their potential beneficial effects on the growth of the host plant (27–31, 39). The suggested mechanisms through which these NREs enhance plant growth include (i) synthesizing plant growth regulators (30, 32, 40–43), (ii) solubilizing phosphate (44), (iii) suppressing plant pathogens (29, 31, 45, 46), and/or (iv) helping the host plant to regulate stresses (46–49). The positive role of these isolated NREs in plant growth was also confirmed by subsequent coinoculations of NRE strains along with rhizobia (27, 28, 30–32, 43, 50). These culture-based studies have provided useful information on the potential beneficial effects of NREs for the host plant. However, culture-based studies cannot provide information on the relative abundance of NREs within root nodules and their preferential selection by host plants, as commonly observed in the case of rhizobial endophytes.
Culture-independent studies have reported that NREs can be highly diverse and abundant (up to 50%) in nodules of soybean (14, 16, 51) and other legume crops (15, 52). However, these studies pooled multiple root nodules per plant or nodules from different plants to assess the overall diversity and abundance of NREs, leaving questions regarding whether the observed diversity and relative abundance are uniformly distributed across all root nodules or whether only a subset of the nodules contain most of the NREs. It is also unclear whether the selection of NRE strains in nodules is a random process, i.e., whichever strain is in close proximity of root nodule formation sites can enter, or if their selection in root nodules is strongly controlled by the host plant to favor a few frequently reported specific NREs based on their beneficial role in the growth of host plant.
Hence, the primary objectives of the present study were (i) to assess the relative distribution and preferential selection of bacterial endophytes within different root nodules of a single plant and (ii) to determine to what extent size or location of root nodules influences the selection of NREs within nodules. We analyzed the microbiomes of 193 individual root nodules from nine soybean plants that were grown under field conditions. We randomly selected 20 to 24 root nodules per plant across the entire root system, and the bacterial endophytes in each were analyzed through 16S rRNA gene amplicon sequencing using Illumina MiSeq paired-end DNA sequencing.
RESULTS AND DISCUSSION
To assess the relative distribution of bacterial endophytes in different root nodules of a plant and in rhizosphere soil, we collected root-adhering soil and root nodules. The rhizosphere and root nodule endophytic bacterial communities were identified through 16S rRNA gene sequencing. The different root nodules varied in size and were collected from different locations of the root system, including the tap and lateral roots (Fig. S1). Overall, we retrieved 3,657,224 bacterial sequences that could be identified at genus level from 193 individual root nodules and 276,338 identifiable sequences from the rhizospheres of nine different soybean plants grown at the Kindrick farm (Greene County, Missouri). The number of good-quality DNA sequences per nodule varied from approximately 13,000 to 55,000. Bradyrhizobium-related sequences were consistently detected in high abundance in almost all root nodules per plant (Fig. 1) as well as across all nine plants (Fig. 2; Table 1). Nitrobacter and Tardiphaga were the two nonrhizobial genera that were detected within nearly all root nodules at low abundance (Fig. 1 and 2; Table 1). Other frequently reported bacterial endophytes were detected at very low abundance (1,635 sequences), and their occurrence was inconsistent within different root nodules of a single plant (Table S2) as well as across different plants (Table 1). Within the rhizobial endophytes, the genus Bradyrhizobium was detected in high abundance (>90% of sequences) in each root nodule. We also detected other soybean-compatible rhizobia in root nodules and rhizosphere, though they were in very low abundance (Fig. 3).
FIG 1.
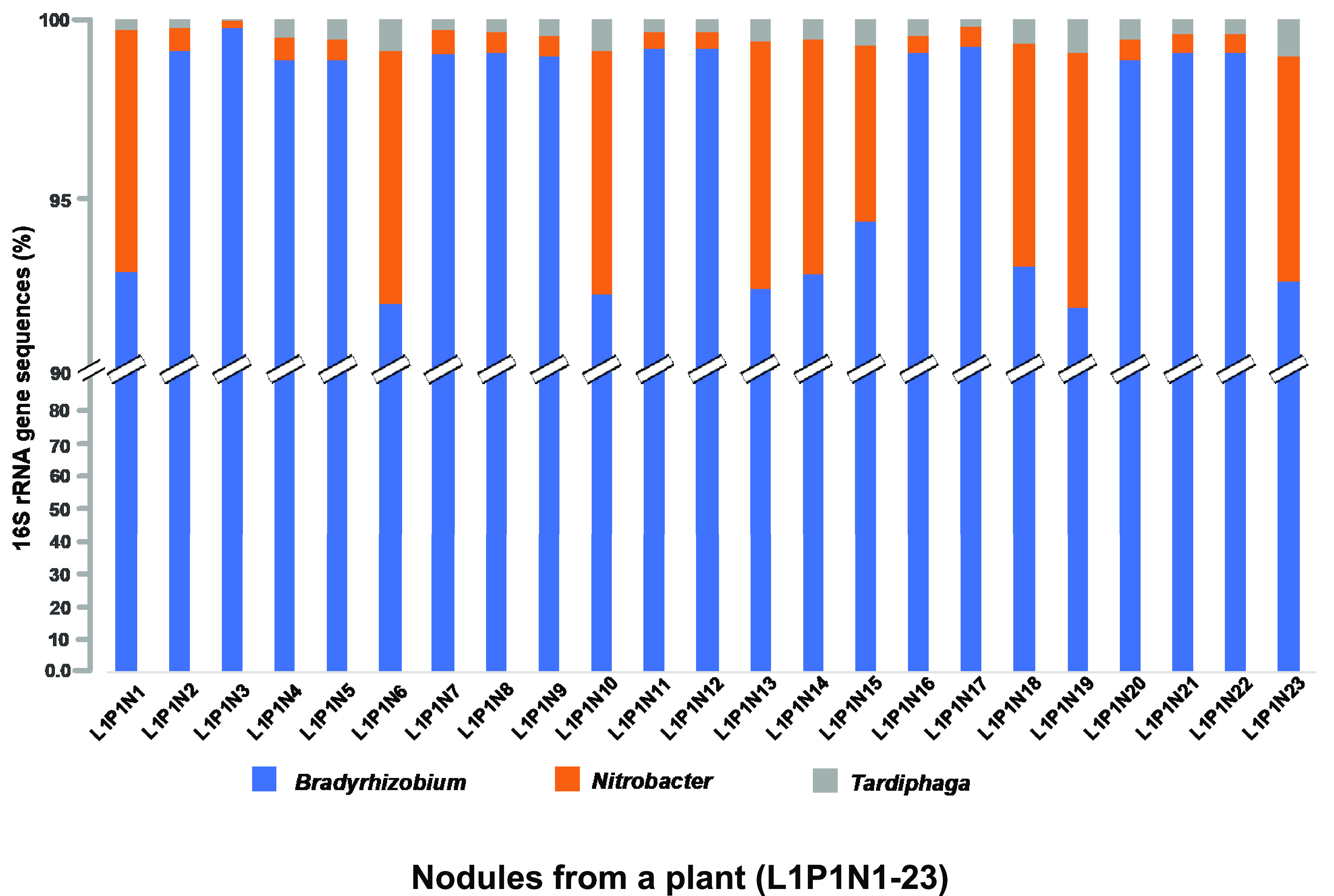
Relative distribution of bacterial endophytes within different root nodules of a soybean plant (L1, P1, N1 to N23) based on 16S rRNA sequence data.
FIG 2.
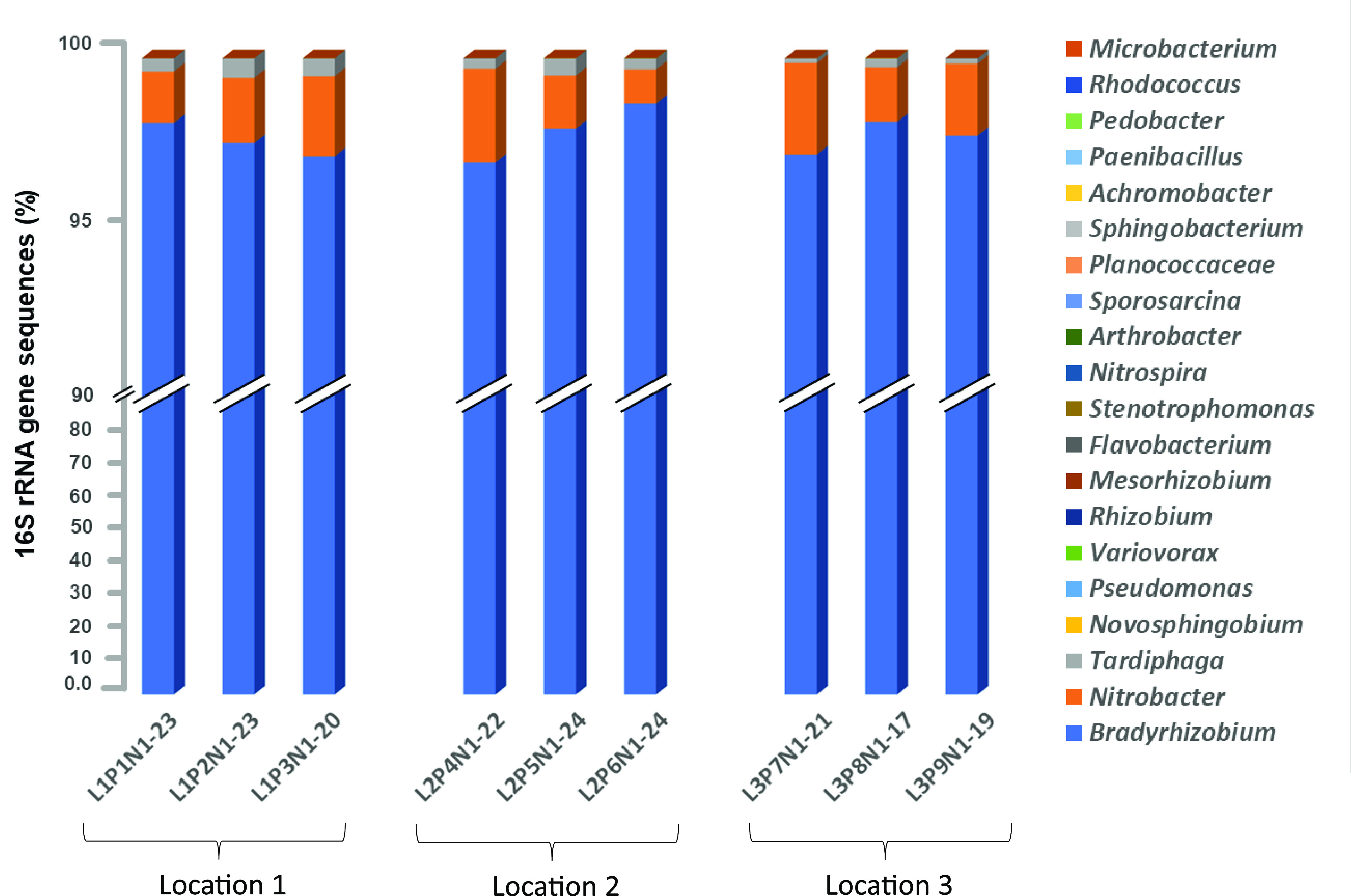
Relative abundance of different bacterial endophytes within different root nodules per plant based on 16S rRNA gene sequencing. The microbiomes of 17 to 24 root nodules per plant were analyzed.
TABLE 1.
Distribution of frequently reported nonrhizobial endophyte sequences in soybean root nodules and rhizosphere soil
| Bacterial genus | No. (% of nodulesa) in: |
|||||
|---|---|---|---|---|---|---|
| Nodule |
Rhizosphere |
|||||
| Location 1 | Location 2 | Location 3 | Location 1 | Location 2 | Location 3 | |
| Novosphingobium | 40 (37) | 52 (27) | 11 (16) | 86 | 77 | 122 |
| Pseudomonas | 11 (9) | 36 (20) | 5 (7) | 6,276 | 10,929 | 15,409 |
| Variovorax | 7 (7) | 20 (12) | 5 (5) | 433 | 889 | 1,513 |
| Paenibacillus | 18 (11) | 3 (1) | 5 (2) | 1,879 | 2944 | 1731 |
| Flavobacterium | 8 (10) | 16 (18) | 3 (4) | 5,989 | 724 | 3,102 |
| Stenotrophomonas | 3 (3) | 2 (2) | 2 (<1) | 1,768 | 246 | 3,602 |
| Nitrospira | 2 (<1) | 2 (<1) | 1 (<1) | 233 | 67 | 550 |
| Arthrobacter | 0 | 4 (2) | 0 | 5,646 | 928 | 3,692 |
| Sporosarcina | 1 (<1) | 3 (2) | 0 | 39,448 | 25,007 | 5,323 |
| Planococcaceae | 0 | 2 (<1) | 1 (<1) | 1,107 | 7,957 | 1,050 |
| Sphingobacterium | 0 | 1 (<1) | 1 (<1) | 7,873 | 183 | 65 |
| Achromobacter | 0 | 1 (<1) | 0 | 301 | 94 | 15,092 |
| Consistently detected bacterial endophytes in almost all root nodules | ||||||
| Bradyrhizobium | 1,499,546 (100) | 1,372,198 (100) | 658,879 (100) | 4,393 | 2,413 | 14,940 |
| Nitrobacter | 42,628 (100) | 36,868 (100) | 24,097 (100) | 112 | 41 | 461 |
| Tardiphaga | 11,109 (98) | 7,925 (97) | 1,808 (97) | 9 | 37 | 48 |
Numbers in parentheses represent the percentage of 193 root nodules that showed the presence of these sequences.
FIG 3.
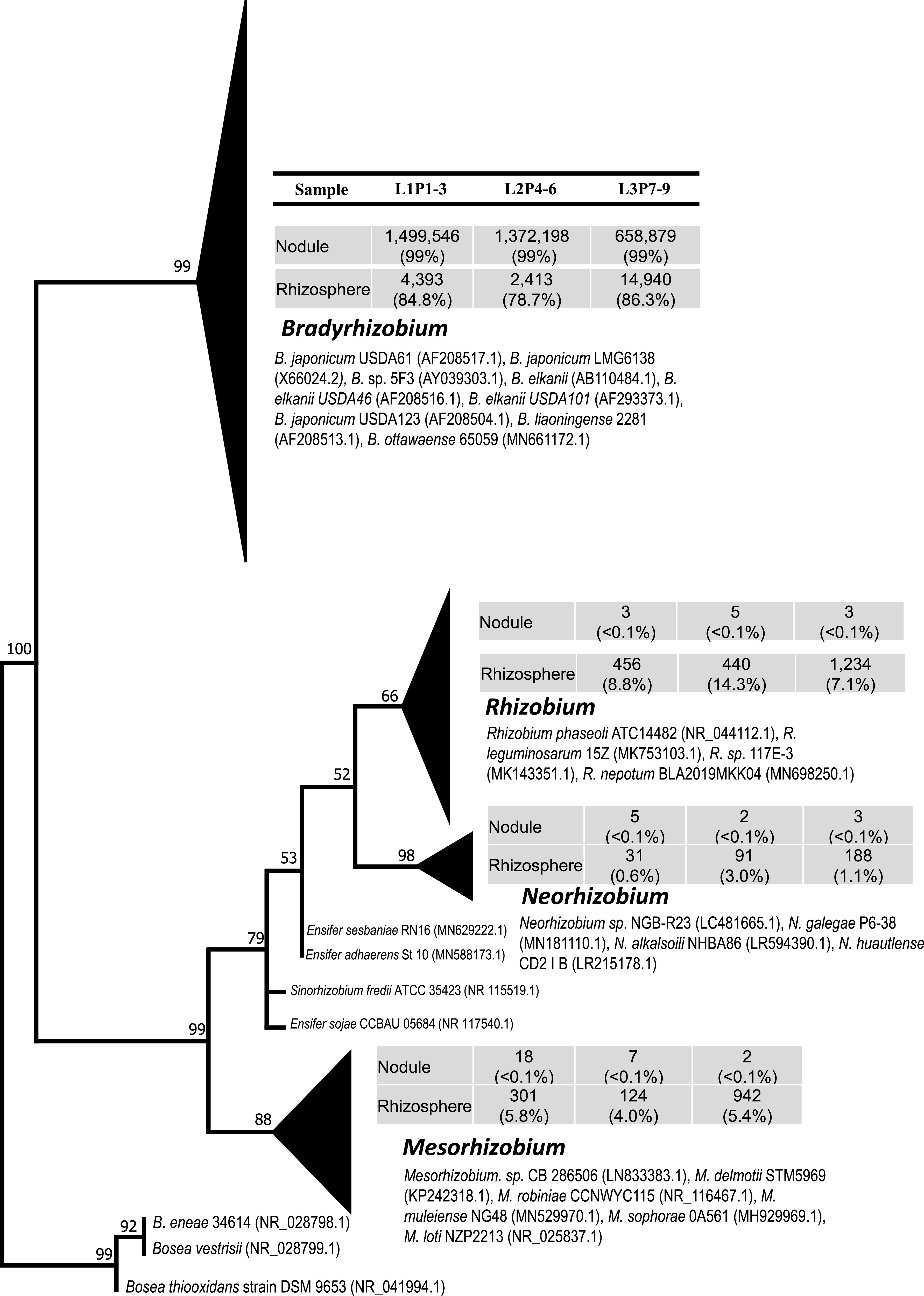
Maximum-likelihood phylogenetic tree based on partial sequences of the 16S rRNA gene of the rhizobial sequences detected within root nodules and rhizosphere of soybean plants. Three locations (L1 to L3) and three plants per location (P1 to P9) were analyzed. Sequences belonging to the same bacterial genera were clustered together. The relative distribution of the total rhizobial sequences retrieved from the root nodules or rhizosphere soils are reported as percentages. The size of the triangle is proportionate to the number of sequences within a genus from nodules and rhizosphere. The GenBank numbers for representative sequences of isolated organisms are shown next to the cluster. The numbers at the nodes reflect bootstrap support values above 50%.
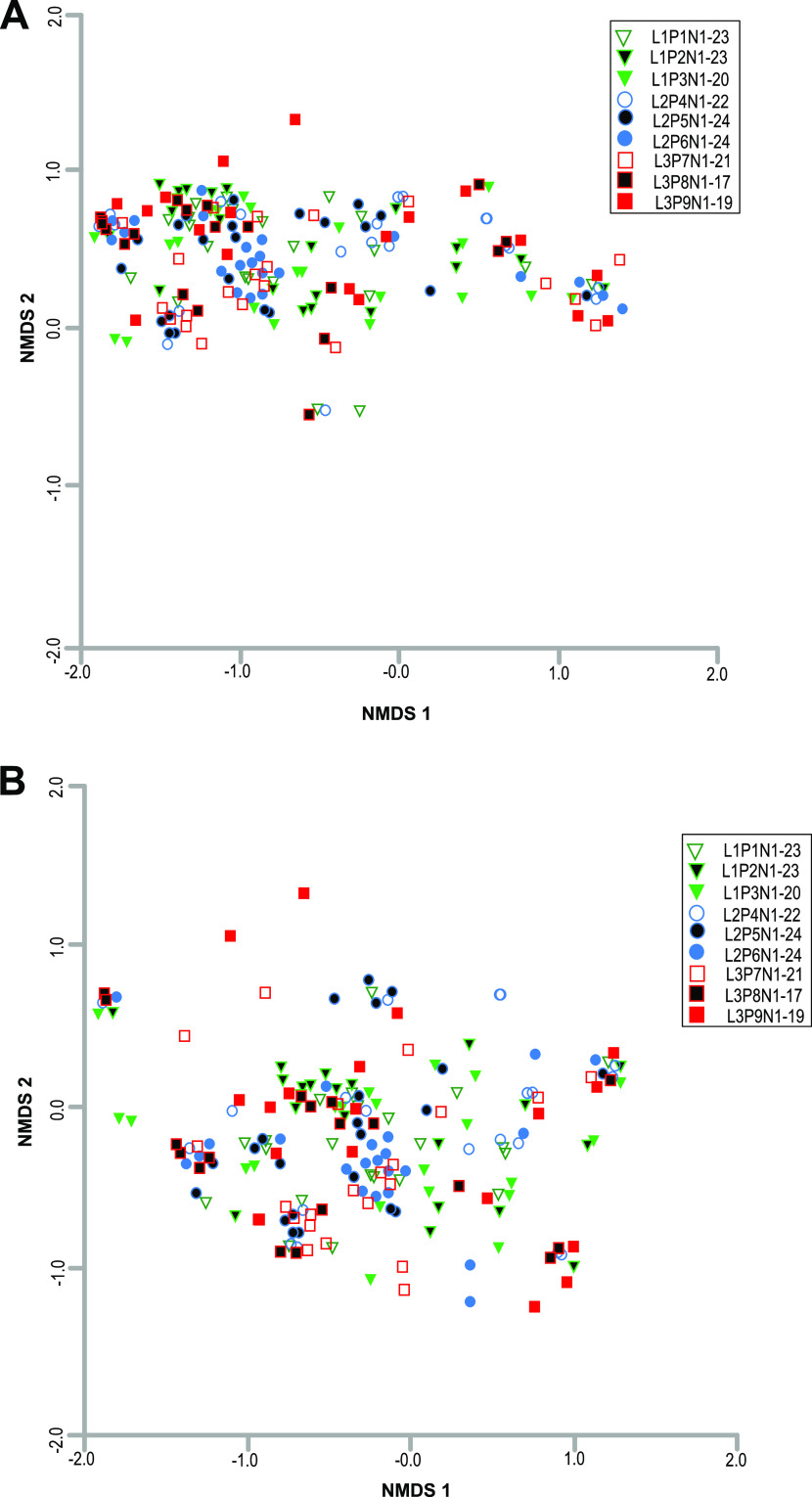
Overall, bacterial community structure in different root nodules, assessed based on Bray-Curtis similarity (97% DNA identity), did not show any significant differences in the clustering of sequences originating from the same plant or same sampling location (permutational multivariate analysis of variance [PERMANOVA], P > 0.05) (Fig. 4A). A similar trend was also observed for other nonrhizobial endophytes after the removal of sequences of three dominant bacterial genera, Bradyrhizobium, Nitrobacter, and Tardiphaga (P > 0.05) (Fig. 4B). However, the greater dispersion in nonmetric multidimensional scaling (NMDS) data points indicated wider variation in the distribution of other inconsistent NREs (Fig. 4B).
FIG 4.
Multidimensional scaling plot, based on the Bray-Curtis similarity index (97% DNA similarity), of the 16S rRNA gene sequences from 193 root nodules of nine plants. Each point represents the microbiome of a single root nodule. Green triangles, P1 to P3 (location 1); blue circles, P4 to P6 (location 2); red squares, P7 to P9 (location 3). The nodules from the different plants from the same location are differentiated by open, color-filled, and black-filled shapes. (A) All bacterial sequences (3,657,224) from both rhizobial and nonrhizobial endophytes were included in this analysis. 2D stress = 0.10. (B) Nonrhizobial endophyte sequences (1,635) excluding the sequences related to Bradyrhizobium, Nitrobacter, and Tardiphaga. These three genera were removed to show the distribution of rare NREs. 2D stress = 0.13.
We evaluated the Chao 1 estimator of species richness and Shannon diversity within nine plants (Table S3). Shannon diversity within root nodules of the different plants ranged from 2.03 to 2.83, which was significantly lower than bacterial diversity in rhizosphere soil (5.72 to 6.40). As anticipated, the root nodule is an enriched locale for certain specific bacteria species, whereas soil is a highly heterogeneous environment which supports diverse bacterial communities (Fig. S2; Table S3). Rarefaction curve analysis indicates that the number of bacterial species within nodules did not reach an asymptote, which suggests that additional sequencing could capture more diversity (Fig. S3). However, rather than overall bacterial diversity, the relative abundance of frequently reported bacterial endophytes within various root nodules of a plant was the main focus of this study to demonstrate the preferential selection of endophytes by host plant.
Distribution of rhizobial endophytes in root nodules and rhizosphere.
As anticipated, the most dominant sequences (93% to 99%) retrieved from each of the 193 root nodules were related to genus Bradyrhizobium (Fig. 1 and 2). Regardless of the location (tap or lateral roots) or size of the root nodules (Fig. S4A and B), the relative abundance of Bradyrhizobium sequences was consistently high (>90%) in all nodules (Fig. 1 and 2; Fig. S2; Table 1). Previous studies (4, 12, 43, 53–56) reported that Bradyrhizobium spp. are major microsymbionts within soybean root nodules. However, members of other rhizobial genera such as Mesorhizobium (6), Rhizobium (7, 8), and Ensifer (9–11) were also reported to nodulate soybean. Interestingly, we observed the presence of DNA sequences of other soybean-compatible rhizobia both within root nodules and in the rhizosphere. However, the relative abundance of other rhizobial endophytes compared to Bradyrhizobium was very low (Fig. 3). This observation suggests a preferred selection of Bradyrhizobium strains by the host plant, which was not influenced by location of the root nodule on the host. Likely, soil surrounding each of 193 root nodules would not be identical in terms of factors such as temperature, O2 concentration, pH, macro- and micronutrients, etc. It is important to mention that we could not measure the geochemical characteristics of soil surrounding each root nodule due to the insufficient amount of soil associated with each root nodule (approximately <0.5 g). For the geochemical analysis, we would need approximately 20 to 30 g of soil. In addition, the collection of soil directly associated with each root nodule was technically not feasible.
Previous studies (10, 53, 57) have reported a high dominance of Ensifer spp. in soybean root nodules in alkaline soils, whereas Bradyrhizobium spp. were detected as major rhizobial endophytes under acidic soil conditions (5). In general, the soils at the study site were highly acidic (pH 5.0 to 5.5), and the pH at this field location was improved through agricultural lime (calcium carbonate) amendment before the sowing of soybean seeds. The final pH of the rhizosphere at the time of plant harvesting (R6 plant growth stage) was slightly acidic (pH 6.5 to 6.8) (Table S4). In the present study, we did not detect any Ensifer-related sequence in 193 nodules, and only 62 Ensifer-related sequences were retrieved from the rhizosphere (Fig. 3).
In the rhizosphere, we observed a relatively high abundance of Bradyrhizobium-related sequences (21,746; 85% of total soil rhizobial sequences) followed by Rhizobium-related sequences (2,130; 8%). Sequences of Mesorhizobium and Neorhizobium were detected in very low abundance (<5%) in the rhizosphere (Fig. 3). Our results suggest that Bradyrhizobium japonicum not only was preferred as the endosymbiont in root nodules but also may have been selectively favored in the rhizosphere. This observation was consistent with the work of Minami et al. (58) and Moawad et al. (59), who suggested that soybean root exudates selectively favored the growth of Bradyrhizobium spp. within the rhizosphere, compared to other rhizobial genera. However, it is important to note that we did not collect bulk soil samples separate from the rhizosphere; therefore, it was not possible to determine whether Bradyrhizobium spp. were selectively enriched in the rhizosphere or were naturally abundant in our soils compared to other rhizobial genera.
We also observed variations in the geochemical characteristics of rhizosphere soils at the three sampling locations. The soil at location 3 had significantly higher concentrations of NO3-N (26.7 ± 1.8 ppm) and K (134 ± 7 mg/liter) than soil the other two locations (Table S4). Location 1 had a higher concentration of NH4-N compared to the other two locations. Location 2 had significantly lower concentrations of phosphorus (P) than the other two locations. Despite the significant variations in soil characteristics (Table S4), we did not observe any specific clustering of endophyte community structure based on the sequences originating from the same plant or same sampling location (Fig. 4; Fig. S4). We observed a selection of B. japonicum strains in root nodules at all three sampling locations. The consistent high abundance of Bradyrhizobium in all 193 root nodules suggests that the influence of specific microenvironments such as different nodules on a plant or nodules across three different locations did not affect the selection of rhizobia. Despite differences in the geochemical characteristics, all three locations were in the same field, and therefore, the climatic conditions were the same. In contrast, previous studies suggested a very strong role of soil characteristics in the selection of rhizobial endophytes within soybean root nodules (4, 6, 9, 21, 22). These previous studies were conducted across diverse geographic regions marked by significant differences in soil pH, moisture content, temperature, precipitation, etc. Our finding on the lack of influence of geochemical characteristics on the selection of bacterial endophytes is based on a limited number of samples that were collected from a single field. Further studies that include amending the soil with various concentrations of macronutrients and transplanting different soybean genotypes could improve our understanding of the potential role of variable nutrient concentrations on the selection of bacterial endophytes.
Distribution of two consistently detected nonrhizobial endophytes (Nitrobacter and Tardiphaga) in various root nodules and rhizosphere samples.
Among the NREs, Nitrobacter and Tardiphaga were detected in almost all individual root nodules (Fig. 1 and 2). Overall, we retrieved 103,707 sequences related to Nitrobacter, and their relative abundance ranged from 1% to 5% within each individual root nodule (Fig. 1). The average abundance of Nitrobacter in the root nodules was <1% of total bacterial sequence; however, a subset of root nodules contained slightly greater levels of Nitrobacter sequences. For example, nine nodules of plant 1 had 3 to 4% Nitrobacter sequences (Fig. 1). This variation did not appear to be related to nodule size, position on root system (tap versus lateral roots), or distance from the soil surface (Fig. 1; Fig. S1 and S4). Tardiphaga-related sequences were detected in 97% of total root nodules with relative densities of 0 to 1%. Both Nitrobacter and Tardiphaga belong to the family Bradyrhizobiaceae and have shown close genetic similarity with the genus Bradyrhizobium based on 16S rRNA (60, 61) as well as five other housekeeping genes, including atpD, dnaK, gyrB, recA, and ropB (61).
We postulate that the consistent presence of Nitrobacter within soybean root nodules may be due to the following reasons. First, Nitrobacter strains are selected within root nodules due to their beneficial role in plant growth. Previously, Ibiene et al. (62) reported that Nitrobacter can solubilize phosphate and have a positive effect on tomato plant growth through inoculation-based studies. Second, the presence of Nitrobacter within soybean root nodules is a parasitic interaction rather than mutualistic. Nitrobacter strains may overcome the host plant defense because of their high genetic similarity to Bradyrhizobium and potentially survive within the root nodules as chemoorganotrophs. Genome sequencing by Starkenburg (60) reported that 41% of genes (1,300 genes) of Nitrobacter strains are identical to those in Bradyrhizobium.
To assess the variation within the Nitrobacter sequences, a maximum-likelihood phylogenetic analysis was conducted. The phylogenetic analysis of all Nitrobacter-related sequences revealed the presence of six major phylogenetic clusters (Fig. S5). Most of the sequences from nodules and rhizosphere samples were grouped into cluster 6 along with other Nitrobacter species sequences from GenBank (Fig. S5).
As with Nitrobacter, we also observed a consistent presence of Tardiphaga-related sequences in almost all root nodules (190 of 193 nodules) (Fig. 2; Table 1). Overall, Tardiphaga-related sequences (20,949) were grouped into three phylogenetic clusters (Fig. S6). Most of the nodule and rhizosphere sequences were related to three reference sequences of Tardiphaga that were isolated by De Meyer et al. (61) from the root nodules of Robinia pseudoacacia (Fig. S6). Previously, Tardiphaga-related strains were isolated from the root nodules of Glycine max (34), R. pseudoacacia (61), and Vavilovia formosa (63) plants. Tardiphaga spp. are genetically similar to the Bradyrhizobium spp. (61) and reported to have nodM and nodT genes (63), which are involved in signaling to the host plant and root nodule formation (64). Moreover, Tardiphaga isolates have been reported to have a role in the N cycle by carrying out dissimilatory nitrate reduction under anaerobic conditions (65).
In the present study, Bradyrhizobium, Nitrobacter, and Tardiphaga were preferred within almost all root nodules, suggesting selection by the host plant or their enhanced ability to penetrate the root nodules. How Nitrobacter and Tardiphaga penetrate and survive within soybean root nodules is an interesting question. We speculate that these bacteria positively affect plant growth, though evidence for this remains to be provided. In all 193 root nodules, Bradyrhizobium-related sequences were the most abundant. Furthermore, none of the 193 root nodules showed high abundance or presence of Nitrobacter and Tardiphaga over Bradyrhizobium. This suggests that Nitrobacter and Tardiphaga enter the root nodules along with Bradyrhizobium and/or that they are not able to overcome the host plant defense in the absence of Bradyrhizobium. The selection, the penetration into host cells or presence within intracellular spaces, and the potential role of these two genera will be the focus of future studies using fluorescence in situ hybridization microscopy and coinoculation-based approaches.
In general, both Nitrobacter- and Tardiphaga-related strains are rarely isolated as plant endophytes in different crops (34, 61–63) (Table S1). In previous culture-based studies on soybean, it was shown that the lack of Nitrobacter may be due to either their slow growth as chemoorganotrophs or lack of NO2 in the growth medium required for growth as chemolithotrophs. Similarly, Tardiphaga species have a growth rate of up to a 10-day doubling time in culture (63). Our study, along with other culture-independent studies, reports the presence of Nitrobacter and Tardiphaga within root nodules of soybean (34) and alfalfa plants (14, 66).
Distribution of inconsistently detected rare nonrhizobial endophytes that were previously reported as endophytes.
One of the major goals of this research was to assess the influence of size and location of root nodules on the preferential selection, diversity, and relative abundance of frequently reported NREs (Table S1) within different root nodules of a single plant. In the last few years, several studies have identified many NREs, such as Pseudomonas (14, 16, 31, 32, 39, 40, 51, 67), Variovorax (33, 40), Novosphingobium (40), Flavobacterium (5, 16), Achromobacter (36, 38, 40), Bacillus (5, 16, 27, 32, 35, 37, 46, 67), etc., in soybean root nodules. These studies focused on the isolation and characterization of these NREs for several plant growth-promoting attributes, such as N fixation, phytohormone production, or potential as biocontrol agents (29–32, 40–43, 45, 46, 68). Most of these studies also demonstrated the beneficial role of the isolated NREs in plant growth through subsequent coinoculation studies (27, 30, 44, 67). In the present study, we detected a small number of the sequences (1,635) related to these frequently reported NREs in soybean root nodules (<1% of total sequences). Their presence was inconsistent among different root nodules of a single plant (Table S2). For example, Novosphingobium-related sequences were retrieved from approximately 25% of the 193 nodule samples, and Pseudomonas-related sequences were found in only 16% of root nodules. This was true for almost all other frequently reported (Table S1) NRE genera (Table 1). The overall low abundance and inconsistent presence of these genera among different root nodules of a single plant suggest that these frequently reported NREs (Table S1) are not preferentially selected as endophytes by host plants and most likely have a very limited role in plant growth as endophytes.
The variation in the NRE sequences detected across different root nodules seems to be the result of a random process rather than due to the influence of the location of the root nodule, as root nodules in close proximity showed the presence of very different NREs and vice versa. For example, Pseudomonas-related sequences were detected within three root nodules of plant 1 (root nodule 2 [N2], N6, and N16), and these nodules were apart from each other (Fig. S1; Table S2). N16 was on a lateral root, and the other two nodules (N2 and N6) were on taproot systems. N3, N4, N5, and N7 were in close proximity to N6, but none of them showed the presence of Pseudomonas-related sequences. Overall, this was true for almost all other NRE-related sequences, implying that their presence was not influenced by the size or location of the root nodules (Fig. S1 and S4A and B; Table S2).
It is important to mention that detection of rare NREs present in different root nodules most likely reflects true sequences rather than potential contamination from DNA extraction or PCR reagents. The following three lines of evidence suggest that the observed NRE sequences are true sequences. (i) None of our negative-control samples showed any amplification for first or second PCRs. (ii) These rare NREs were detected in only a few nodule samples (Table 1); if these sequences had originated from contaminating reagents, they would be consistently present across all root nodule samples. (iii) We did detect a high abundance of the identical sequences from rhizosphere soil samples as well. Specifically, more than 25,000 Pseudomonas sequences retrieved from rhizosphere soil were identical to 40 Pseudomonas sequences detected in various root nodules. This was also true for nearly all other rare NREs. Furthermore, we only used the DNA sequences that can be classified at a >80% confidence cutoff level, indicating that they are most likely true sequences rather than sequencing artifacts.
Almost all of the prokaryotic sequences that we detected in root nodules were also found in the 276,338 sequences from nine rhizosphere samples. In contrast to root nodules, there was a higher abundance (2% to 25% of total rhizosphere sequences) of the commonly reported NREs, such as Pseudomonas (11.8% of total rhizosphere sequences), Achromobacter (5.6%), Arthrobacter (3.7%), Planococcaceae (3.5%), Flavobacterium (3.5%), Sphingobacterium (2.9%), Paenibacillus (2.4%), and Stenotrophomonas (2%), in the rhizosphere (Table 1). Based on reports of the positive influence of these NREs on the growth of the host plant, these NREs likely have a significant role in plant growth as free-living plant growth-promoting rhizobacteria (27, 28, 30–32, 43).
In the present study, we have carefully surface-sterilized each root nodule and removed the epidermis. The rare NRE-related sequences (excluding Nitrobacter and Tardiphaga sequences) could originate from surface contamination of root nodules. Even though bacterial cells on the root nodule surface may not be culturable on nutrient agar, their DNA can be amplified through PCR. The surface contamination effect could be more pronounced when multiple root nodules from different plants are pooled for the DNA extraction (14, 16). Likewise, frequent isolation of NREs by culture-based studies could be due to their copious growth on a nutrient-rich growth medium, even if they are naturally present in low abundance in root nodules. Therefore, detecting these commonly reported NREs does not necessarily indicate that they play a significant role in plant growth as endophytes.
In summary, we observed a high abundance of Bradyrhizobium species within 193 root nodules of nine plants (>90% sequences within each root nodule). The two nonrhizobial endophytes (Nitrobacter and Tardiphaga) were also consistently detected within almost all root nodules, though they were in low abundance (1 to 3%) compared to Bradyrhizobium. The consistent presence of these three bacterial genera (one rhizobial genus and two NREs) within almost all root nodules suggests their preferential selection by the host plant that was not influenced by the size, age, or location of the root nodules on a root system. In contrast, we detected low frequency and inconsistent presence of other frequently reported soybean NREs (Table S1) such as Bacillus, Pseudomonas, Flavobacterium, and Variovorax species within very few nodules. In other words, no single NRE that was frequently reported as a soybean endophyte in previous studies (Table S1) was detected consistently or in high abundance in most of the root nodules The overall low abundance and inconsistent presence of frequently reported soybean NREs (Table S1) among different root nodules of a single plant suggest that these frequently reported NREs are not preferentially selected as endophytes by host plants and most likely have a very limited role in plant growth as endophytes. Simple isolation and characterization of frequently reported NREs from root nodules alone may not be sufficient to establish their roles as beneficial endophytes. Their relative abundance within root nodules and preferential selection by the host plant are equally important.
MATERIALS AND METHODS
Research site.
Soybean seeds were obtained from Hancock Seed Co., Dade, FL, USA. The seeds were grown at the Kindrick Farm, Green County, Missouri (latitude, 37°21′44.28″N; longitude, −93°40′91.96″W). Seeds were planted in May 2018, and plants were harvested in September 2018 for the microbiome analysis. We harvested plants at the R6 plant growth stage. Soybean seeds were planted with no tillage at row widths of 15 in. Seeds were planted with planter’s disks, which made small slits for seed placement. The no-tillage approach was adopted to reduce disturbance and soil erosion and to increase available moisture compared to conventionally tilled systems. Seeds were not inoculated with any bacterial culture. Seeds were treated with Acceleron standard seed applied solutions (69). No irrigation was applied, and rainwater was the sole source of water for the crop. The total rainfall from May to September 2018 was 19 in. Rainfall averaged 3.8 in. per month and was lowest during July (1.04 in.) and highest during August (6.71 in.). The average monthly temperature for the 5 months was 25°C ± 2°C. Kindrick Farm has a history of corn cropping for the past 7 years. Three locations within the farm were sampled: location 1 (toe slope), location 2 (sinkhole 1), and location 3 (sinkhole 2). The three soil locations and their geochemical characteristics are described in detail in the supplemental material (Fig. S7; Table S4).
Sample collection.
Three replicate plants from three separate rows at each location were excavated, and intact root systems containing root nodules and rhizosphere were brought to the laboratory. We selected all plants of the approximately same height and the same age. Rhizosphere soil was separated from the root systems by gently shaking the plant roots on a sterilized bag. Soil detached in this way was used for geochemical analysis, and soil that remained attached to roots after gentle shaking was used for DNA extraction. For the microbial analysis, root-adhering soil of each plant was washed into separate beakers containing water. The resulting soil suspension was centrifuged at 14,000 rpm for 5 min, and 0.5 g of soil pellet was used for DNA extraction. Roots were then thoroughly washed with distilled water and photographed. Approximately 20 to 24 individual root nodules per plant were selected across the entire root system (Fig. S1). The samples were labeled L1 to L3 for locations, P1 to P9 for plants, and N1 to N24 for nodules per plant (Table S3). We collected the root nodules from both taproots and lateral roots. A taproot, or primary root, is a thick large central root from which lateral roots sprout. The root nodules on the taproot could easily be distinguished from the ones on lateral roots (Fig. S1). A total of 193 individual root nodules from these nine plants were collected for microbiome analysis. Each individual nodule was thoroughly washed in distilled water and surface sterilized in 75% ethanol. The root nodule epidermis was aseptically removed using sterilized forceps while keeping roots submerged in 75% ethanol (Fig. S8). Surface-cleaned root nodules were crushed individually in 1 ml of sterile water using a sterilized mortar and pestle. Crushed root nodule material was used for DNA extraction. Complete surface sterilization of the nodule was confirmed by rolling a few randomly selected nodules on tryptic soy agar plates.
DNA extraction.
DNA was extracted from each root nodule as well as from the rhizosphere samples using the DNeasy PowerLyzer PowerSoil DNA extraction kit (Qiagen, MoBio, USA). Briefly, the crushed root nodule material was centrifuged at 10,000 rpm for 10 min, and the supernatant was discarded. The pellet containing bacterial cells and plant material was suspended in the Power bead solution (provided with the DNA extraction kit), and DNA was extracted according to the manufacturer’s instructions. For soil samples, 0.4 g of rhizosphere soil was used for DNA extraction. Extracted DNA was eluted in 25 μl of sterilized water and stored at −20°C until used in PCR.
Sequencing preparation.
To assess the bacterial communities in the root nodules and rhizosphere, the Illumina MiSeq paired-end DNA sequencing approach was used. Briefly, we used a two-PCR approach. During the first PCR, the partial bacterial 16S rRNA gene fragment of the V3–V5 region was amplified using primers F515 (5′-GTGCCAGCMGCCGCGG-3′) and R907 (5′-CCGTCAATTCMTTTRAGTTT-3′). These universal bacterial primers also contained the sequences for Illumina DNA sequencing primers (Fig. S9). PCR was performed in a 25-μl PCR mixture containing 1× buffer, a 0.2 μM concentration of each primer, 2.0 mM MgSO4, a 0.2 μM concentration of each deoxynucleoside triphosphate (dNTP), 1.0 μl of template, and 1.0 unit of high-fidelity Platinum Taq polymerase (Invitrogen, USA). The PCR conditions were as follows: 3 min at 96°C, followed by 30 cycles of denaturation at 94°C for 45 s, primer annealing at 56°C for 45 s, extension at 72°C for 45 s, and a final extension at 72°C for 7 min. As a standard PCR procedure, we ran positive and negative controls along with each set of PCRs. Amplified PCR products along with the negative controls were cleaned using the ExoSAP-IT PCR cleanup system (Invitrogen, USA), according to the manufacturer’s instructions.
Cleaned PCR products from the first PCR along with the negative controls were diluted (10 times) in PCR-grade water and used as the template in the second PCR. All the reagents used in the second PCR were as described above with the exception of PCR primers. The primers used in the second PCR contained the Illumina sequencing adapters A and B (A, 5′-CAAGCAGAAGACGGCATACGAGAT-3′; B, 5′-AATGATACGGCGACCACCGAGATCTACA-3′) and standard unique multiplex identifier sequences (listed in Table S5) for each sample (Fig. S9; Table S5). The PCR conditions for this second PCR were as follows: initial denaturation for 3 min at 90°C, followed by 10 cycles of denaturation at 94°C for 30 s, annealing at 60°C for 30 s, and extension at 72°C for 30 s, with a final extension step at 72°C for 7 min. PCR-amplified uniquely indexed PCR products were pooled in equimolar concentrations. Pooled, amplified DNA products were purified with an Agencourt AMPure magnetic bead system (Beckman Coulter, Brea, CA). Purified PCR products were sequenced using the Illumina MiSeq paired-end DNA sequencing platform.
Data analysis.
Paired-end Illumina reads were assembled to generate consensus sequences, and initial quality filtration was done using mothur software (70). Initial quality filtrations such as min-length = 370, max-ambig = 3, max-homp = 8, etc., were performed as described previously (71). Chimeric DNA sequences were identified and removed using the Ribosomal Database Project (RDP) platform. Plant-associated cyanobacterium/chloroplast sequences were also identified and removed. High-quality DNA sequences (3,657,224) were classified using the RDP naive Bayesian classifier 2.5 (72). These classified DNA sequences were also aligned and clustered at 97% DNA similarity for multivariate analysis.
DNA sequences related to rhizobial genera were retrieved from root nodules (3,530,886 sequences) and rhizosphere (276,338), and were extracted using mothur. The rhizobial sequences along with the reference sequences from GenBank were aligned and clustered into different groups at 97% DNA similarity. The representative sequences from each cluster were selected for the maximum-likelihood (ML) phylogenetic analysis. The phylogenetic analysis was performed to assess the distribution of rhizobial genera within soybean root nodules and rhizosphere. MEGA software version 5.2 (73) was used to construct an ML phylogenetic tree, and branches representing the same genera were collapsed into subgroups. For the phylogenetic tree, the Tamura-Nei model of uniform nucleotide substitution rate was selected, and confidence in tree topology was assessed by 10,000 bootstrap iterations. Bootstrap values greater than 50% were reported in the phylogenetic tree. Similarly, phylogenetic trees for Nitrobacter and Tardiphaga were also constructed.
The variations in the endophytic bacterial community in soybean root nodules of different plants were accessed by nonmetric multidimensional scaling (NMDS) as described earlier (74). Briefly, we used two different data sets for NMDS analysis. The first data set contained the distribution of all classified bacterial genera in root nodules (containing 3,657,224 sequences), whereas the second data set contained only the distribution of 1,635 sequences of NREs, excluding the sequences related to Bradyrhizobium, Nitrobacter, and Tardiphaga. Within almost all root nodules, these three bacterial genera were consistently detected at a relatively high abundance compared to other commonly reported NREs. For both data sets, we have used the abundance-based Bray-Curtis similarity metric, and data were square root transformed prior to calculating the resemblance metric. The Bray-Curtis similarity metric was calculated using the function vegdist in the R package vegan (75), and the significance of differences across different plants as well as at different locations was tested by PERMANOVA (76) using the function adonis in R (75).
Soil analysis.
Briefly, the rhizosphere soils of each plant (P1 to P9) were collected separately by gently shaking the plant roots on a sterilized bag. Approximately 20 to 30 g of soil per plant was collected and sent to the University of Missouri Soil and Plant Testing Laboratory for geochemical analysis. The soil samples were tested for pH, organic matter, nitrate concentration, ammonium concentration, phosphorus, potassium, calcium, magnesium, and potential cation exchange capacity using the procedures described in reference 77.
Data availability.
The 16S rRNA gene sequences were deposited in the NCBI Sequence Read Archive (SRA) under BioProject ID PRJNA625756.
Supplementary Material
ACKNOWLEDGMENTS
We are thankful to Michael G. Burton for providing access to the Kindrick family farm and all his support for soybean crop management-related work.
The funding for this research work was supported by Missouri State University (faculty research grant).
Author contributions: B.S.M. conceived the idea and designed research, P.M. and B.S.M. performed research work and analyzed data, and B.S.M. wrote and revised the manuscript.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Vinuesa P, Rojas-Jiménez K, Contreras-Moreira B, Mahna SK, Prasad BN, Moe H, Selvaraju SB, Thierfelder H, Werner D. 2008. Multilocus sequence analysis for assessment of the biogeography and evolutionary genetics of four Bradyrhizobium species that nodulate soybeans on the Asiatic continent. Appl Environ Microbiol 74:6987–6996. 10.1128/AEM.00875-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chibeba AM, Kyei-Boahen S, de Fátima Guimarães M, Nogueira MA, Hungria M. 2017. Isolation, characterization and selection of indigenous Bradyrhizobium strains with outstanding symbiotic performance to increase soybean yields in Mozambique. Agric Ecosyst Environ 246:291–305. 10.1016/j.agee.2017.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Han LL, Wang ET, Han TX, Liu J, Sui XH, Chen WF, Chen WX. 2009. Unique community structure and biogeography of soybean rhizobia in the saline-alkaline soils of Xinjiang. Plant Soil 324:291–305. 10.1007/s11104-009-9956-6. [DOI] [Google Scholar]
- 4.Shiro S, Matsuura S, Saiki R, Sigua GC, Yamamoto A, Umehara Y, Hayashi M, Saeki Y. 2013. Genetic diversity and geographic distribution of indigenous soybean-nodulating bradyrhizobia in the United States. Appl Environ Microbiol 79:3610–3618. 10.1128/AEM.00236-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang YM, Li Y, Jr, Chen WF, Wang ET, Tian CF, Li QQ, Zhang YZ, Sui XH, Chen WX. 2011. Biodiversity and biogeography of rhizobia associated with soybean plants grown in the north China plain. Appl Environ Microbiol 77:6331–6342. 10.1128/AEM.00542-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Adhikari D, Kaneto M, Itoh K, Suyama K, Pokharel BB, Gaihre YK. 2012. Genetic diversity of soybean-nodulating rhizobia in Nepal in relation to climate and soil properties. Plant Soil 357:131–145. 10.1007/s11104-012-1134-6. [DOI] [Google Scholar]
- 7.Kapembwa R, Mweetwa AM, Ngulube M, Yengwe J, Yengwe J. 2016. Morphological and biochemical characterization of soybean nodulating rhizobia indigenous to Zambia. Sustain Agric Res 5:84–92. 10.5539/sar.v5n3p84. [DOI] [Google Scholar]
- 8.Scholla MH, Elkan HG. 1984. Rhizobium fredii sp.nov. a fast-growing species that effectively nodulates soybeans. Int J Syst Bacteriol 34:484–486. 10.1099/00207713-34-4-484. [DOI] [Google Scholar]
- 9.Yan J, Han XZ, Ji ZH, Li Y, Wang ET, Xie ZH, Chen WF. 2014. Abundance and diversity of soybean-nodulating rhizobia in black soil are impacted by land use and crop management. Appl Environ Microbiol 80:5394–5402. 10.1128/AEM.01135-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pastorino G, Alcántara V, Malbrán I, Videira L, Sarinelli J, Balatti P. 2015. Ensifer (Sinorhizobium) fredii interacted more efficiently than Bradyrhizobium japonicum with soybean. J Agric Ecol Res Int 2:10–19. 10.9734/JAERI/2015/13163. [DOI] [Google Scholar]
- 11.Peng GX, Tan ZY, Wang ET, Reinhold-Hurek B, Chen WF, Chen WX. 2002. Identification of isolates from soybean nodules in Xinjiang region as Sinorhizobium xinjiangense and genetic differentiation of S. xinjiangense from Sinorhizobium fredii. Int J Syst Evol Microbiol 52:457–462. 10.1099/00207713-52-2-457. [DOI] [PubMed] [Google Scholar]
- 12.Saeki Y, Minami M, Yamamoto A, Akao S. 2008. Estimation of the bacterial community diversity of soybean-nodulating rhizobia isolated from Rj-genotype soybean. Soil Sci Plant Nutr 54:718–724. 10.1111/j.1747-0765.2008.00300.x. [DOI] [Google Scholar]
- 13.Zahran HH. 1999. Rhizobium-legume symbiosis and nitrogen fixation under severe conditions and in an arid climate. Microbiol Mol Biol Rev 63:968–989. 10.1128/MMBR.63.4.968-989.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xiao X, Chen W, Zong L, Yang J, Jiao S, Lin Y, Wang E, Wei G. 2017. Two cultivated legume plants reveal the enrichment process of the microbiome in the rhizocompartments. Mol Ecol 26:1641–1651. 10.1111/mec.14027. [DOI] [PubMed] [Google Scholar]
- 15.Wigley K, Moot D, Wakelin SA, Laugraud A, Blond C, Seth K, Ridgway H. 2017. Diverse bacterial taxa inhabit root nodules of lucerne (Medicago sativa L.) in New Zealand pastoral soils. Plant Soil 420:253–262. 10.1007/s11104-017-3395-6. [DOI] [Google Scholar]
- 16.Sharaf H, Rodrigues RR, Moon J, Zhang B, Mills K, Williams MA. 2019. Unprecedented bacterial community richness in soybean nodules vary with cultivar and water status. Microbiome 7:63. 10.1186/s40168-019-0676-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Al-Falih AMK. 2002. Factors affecting the efficiency of symbiotic nitrogen fixation by Rhizobium. Pakistan J Biol Sci 5:1277–1293. 10.3923/pjbs.2002.1277.1293. [DOI] [Google Scholar]
- 18.Mason MLT, Tabing BLC, Yamamoto A, Saeki Y. 2018. Influence of flooding and soil properties on the genetic diversity and distribution of indigenous soybean-nodulating Bradyrhizobia in the Philippines. Heliyon 4:e00921. 10.1016/j.heliyon.2018.e00921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thies JE, Singleton PW, Bohlool BB. 1991. Influence of the size of indigenous rhizobial populations on establishment and symbiotic performance of introduced rhizobia on field-grown legumes. Appl Environ Microbiol 57:19–28. 10.1128/AEM.57.1.19-28.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Singleton PW, Bohlool BB, Nakao PL. 1992. Legume response to rhizobial inoculation in the tropics: myths and realities, p 135–155. In Lal R, Sanchez PA (ed), Myths and science of soils of the tropics, vol 29. SSSA Publications, Madison, WI. [Google Scholar]
- 21.Herridge DF, Roughley RJ, Brockwell J. 1984. Effect of rhizobia and soil nitrate on the establishment and functioning of the soybean symbiosis in the field. Aust J Agric Res 35:149–161. 10.1071/AR9840149. [DOI] [Google Scholar]
- 22.Wasike VW, Lesueur D, Wachira FN, Mungai NW, Mumera LM, Sanginga N, Mburu HN, Mugadi D, Wango P, Vanlauwe B. 2009. Genetic diversity of indigenous Bradyrhizobium nodulating promiscuous soybean Glycine max (L.) Merr. varieties in Kenya: impact of phosphorus and lime fertilization in two contrasting sites. Plant Soil 322:151–163. 10.1007/s11104-009-9902-7. [DOI] [Google Scholar]
- 23.Thies JE, Bohlool BB, Singleton PW. 1992. Environmental effects on competition for nodule occupancy between introduced and indigenous rhizobia and among introduced strains. Can J Microbiol 38:493–500. 10.1139/m92-081. [DOI] [Google Scholar]
- 24.Osunde A, Gwam S, Bala A, Sanginga N, Okogun J. 2003. Responses to rhizobial inoculation by two promiscuous soybean cultivars in soils of the Southern Guinea savanna zone of Nigeria. Biol Fertil Soils 37:274–279. 10.1007/s00374-003-0609-2. [DOI] [Google Scholar]
- 25.O'Brien SL, Gibbons SM, Owens SM, Hampton-Marcell J, Johnston ER, Jastrow JD, Gilbert JA, Meyer F, Antonopoulos DA. 2016. Spatial scale drives patterns in soil bacterial diversity. Environ Microbiol 18:2039–2051. 10.1111/1462-2920.13231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Štursová M, Bárta J, Šantrůčková H, Baldrian P. 2016. Small-scale spatial heterogeneity of ecosystem properties, microbial community composition and microbial activities in a temperate mountain forest soil. FEMS Microbiol Ecol 92:fiw185. 10.1093/femsec/fiw185. [DOI] [PubMed] [Google Scholar]
- 27.Bai Y, D'Aoust F, Smith DL, Driscoll BT. 2002. Isolation of plant-growth-promoting Bacillus strains from soybean root nodules. Can J Microbiol 48:230–238. 10.1139/w02-014. [DOI] [PubMed] [Google Scholar]
- 28.Dashti N, Zhang F, Hynes R, Smith DL. 1997. Application of plant growth-promoting rhizobacteria to soybean [Glycine max (L.) Merr] increases protein and dry matter yield under short-season conditions. Plant Soil 188:33–41. 10.1023/A:1004295827311. [DOI] [Google Scholar]
- 29.Liu ZL, Sinclair JB. 1993. Colonization of soybean roots by Bacillus megaterium B153-2–2. Soil Biol Biochem 25:849–855. 10.1016/0038-0717(93)90087-R. [DOI] [Google Scholar]
- 30.Mishra PK, Mishra S, Selvakumar G, Kundu S, Gupta HS. 2009. Enhanced soybean (Glycine max L.) plant growth and nodulation by Bradyrhizobium japonicum-SB1 in presence of Bacilllus thuringiensis-KR1. Acta Agric Scand B Soil Plant Sci 59:189–196. 10.1080/09064710802040558. [DOI] [Google Scholar]
- 31.Tokgöz S. 2018. Exploration of soybean nodule microbiome for plant health management. MS thesis. University of Nebraska, Lincoln, NE. [Google Scholar]
- 32.Zhao L, Xu Y, Lai X. 2018. Antagonistic endophytic bacteria associated with nodules of soybean (Glycine max L.) and plant growth promoting properties. Braz J Microbiol 49:269–278. 10.1016/j.bjm.2017.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.de Almeida Lopes KB, Carpentieri-Pipolo V, Oro TH, Stefani-Pagliosa E, Degrassi G. 2016. Culturable endophytic bacterial communities associated with field-grown soybean. J Appl Microbiol 120:740–755. 10.1111/jam.13046. [DOI] [PubMed] [Google Scholar]
- 34.Bromfield ESP, Cloutier S, Tambong JT, Tran TT. 2017. Soybeans inoculated with root zone soils of Canadian native legumes harbour diverse and novel Bradyrhizobium spp. that possess agricultural potential. Syst Appl Microbiol 40:440–447. 10.1016/j.syapm.2017.07.007. [DOI] [PubMed] [Google Scholar]
- 35.Zhang YZ, Chen WF, Li M, Sui XH, Liu H-C, Zhang XX, Chen WX. 2012. Bacillus endoradicis sp. nov., an endophytic bacterium isolated from soybean root. Int J Syst Evol Microbiol 62:359–363. 10.1099/ijs.0.028936-0. [DOI] [PubMed] [Google Scholar]
- 36.Wedhastri S, Fardhani DM, Kabirun S, Widada J, Widianto D, Evizal R, Prijambada ID. 2015. Legume nodulating bacterium, Achromobacter xylosoxidans found in tropical shrub agroecosystem. Indones J Biotechnol 18:161–167. 10.22146/ijbiotech.7879. [DOI] [Google Scholar]
- 37.Annapurna K, Ramadoss D, Bose P, Vithalkumar L. 2013. In situ localization of Paenibacillus polymyxa HKA-15 in roots and root nodules of soybean (Glycine max L.). Plant Soil 373:641–648. 10.1007/s11104-013-1825-7. [DOI] [Google Scholar]
- 38.Hung PQ, Kumar SM, Govindsamy V, Annapurna K. 2007. Isolation and characterization of endophytic bacteria from wild and cultivated soybean varieties. Biol Fertil Soils 44:155–162. 10.1007/s00374-007-0189-7. [DOI] [Google Scholar]
- 39.Kuklinsky-Sobral J, Araujo WL, Mendes R, Geraldi IO, Pizzirani-Kleiner AA, Azevedo JL. 2004. Isolation and characterization of soybean-associated bacteria and their potential for plant growth promotion. Environ Microbiol 6:1244–1251. 10.1111/j.1462-2920.2004.00658.x. [DOI] [PubMed] [Google Scholar]
- 40.Aserse AA, Rasanen LA, Aseffa F, Hailemariam A, Lindstrom K. 2013. Diversity of sporadic symbionts and nonsymbiotic endophytic bacteria isolated from nodules of woody, shrub, and food legumes in Ethiopia. Appl Microbiol Biotechnol 97:10117–10134. 10.1007/s00253-013-5248-4. [DOI] [PubMed] [Google Scholar]
- 41.Martínez-Hidalgo P, Hirsch AM. 2017. The nodule microbiome: N2 -fixing Rhizobia do not live alone. Phytobiomes J 1:70–82. 10.1094/PBIOMES-12-16-0019-RVW. [DOI] [Google Scholar]
- 42.Mirza BS, Mirza MS, Bano A, Malik KA. 2007. Co-inoculation of chickpea with Rhizobium isolates from roots and nodules and phytohormone-producing Enterobacter strains. Aust J Exp Agric 47:1008. 10.1071/EA06151. [DOI] [Google Scholar]
- 43.Masciarelli O, Llanes A, Luna V. 2014. A new PGPR co-inoculated with Bradyrhizobium japonicum enhances soybean nodulation. Microbiol Res 169:609–615. 10.1016/j.micres.2013.10.001. [DOI] [PubMed] [Google Scholar]
- 44.Li JH, Wang E, Chen WF, Chen XC. 2008. Genetic diversity and potential for promotion of plant growth detected in nodule endophytic bacteria of soybean grown in Heilongjiang Province of China. Soil Biol Biochem 40:238–246. 10.1016/j.soilbio.2007.08.014. [DOI] [Google Scholar]
- 45.Le Cocq K, Gurr SJ, Hirsch PR, Mauchline TH. 2017. Exploitation of endophytes for sustainable agricultural intensification. Mol Plant Pathol 18:469–473. 10.1111/mpp.12483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Senthilkumar M, Swarnalakshmi K, Gov-Indasamy V, Lee YK, Annapurna K. 2009. Biocontrol potential of soybean bacterial endophytes against charcoal rot fungus, Rhizoctonia bataticola. Curr Microbiol 58:288–293. 10.1007/s00284-008-9329-z. [DOI] [PubMed] [Google Scholar]
- 47.Dhole A, Shelat H. 2018. Non-rhizobial endophytes in root nodules. MOJ Biol Med 3:147–148. 10.15406/mojbm.2018.03.00064. [DOI] [Google Scholar]
- 48.Yang J, Kloepper JW, Ryu CM. 2009. Rhizosphere bacteria help plants tolerate abiotic stress. Trends Plant Sci 14:1–4. 10.1016/j.tplants.2008.10.004. [DOI] [PubMed] [Google Scholar]
- 49.Han HS, Lee KD. 2005. Physiological responses of soybean—inoculation of Bradyrhizobium japonicum with PGPR in saline soil conditions. Res J Agric Biol Sci 1:216–221. [Google Scholar]
- 50.Tariq M, Hameed S, Yasmeen T, Ali A. 2012. Non-rhizobial bacteria for improved nodulation and grain yield of mung bean [Vigna radiata (L.) Wilczek]. Afr J Biotechnol 11:15012–15019. [Google Scholar]
- 51.Zhang B, Du N, Li Y, Shi P, Wei G. 2018. Distinct biogeographic patterns of rhizobia and non-rhizobial endophytes associated with soybean nodules across China. Sci Total Environ 643:569–578. 10.1016/j.scitotenv.2018.06.240. [DOI] [PubMed] [Google Scholar]
- 52.Lu J, Yang F, Wang S, Ma H, Liang J, Chen Y. 2017. Co-existence of rhizobia and diverse non-rhizobial bacteria in the rhizosphere and nodules of Dalbergia odorifera seedlings inoculated with Bradyrhizobium elkanii, rhizobium multihospitium-like and Burkholderia pyrrocinia-like strains. Front Microbiol 8:2255. 10.3389/fmicb.2017.02255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Saeki Y, Kaneko A, Hara T, Suzuki K, Yamakawa T, Nguyen MT, Nagatomo Y, Akao S. 2005. Phylogenetic analysis of soybean-nodulating rhizobia isolated from alkaline soils in Vietnam. Soil Sci Plant Nutr 51:1043–1052. 10.1111/j.1747-0765.2005.tb00143.x. [DOI] [Google Scholar]
- 54.Kuykendall LD, Saxena B, Devine TE, Udell SE. 1992. Genetic diversity in Bradyrhizobium japonicum Jordan 1982 and a proposal for Bradyrhizobium elkanii sp. nov. Can J Microbiol 38:501–505. 10.1139/m92-082. [DOI] [Google Scholar]
- 55.Xu ML, Ge C, Cui Z, Li J, Fan H. 1995. Bradyhizobium liaoningense sp. nov., isolated from the root nodules of soybeans. Int J Syst Bacteriol 45:706–711. 10.1099/00207713-45-4-706. [DOI] [PubMed] [Google Scholar]
- 56.Yao YZ, Kan FL, Wang ET, Wei GH, Chen X. 2002. Characterization of rhizobia that nodulate legume species of the genus Lespedeza and description of Bradyrhizobium yuanmingense sp. nov. Int J Syst Evol Microbiol 52:2219–2230. 10.1099/00207713-52-6-2219. [DOI] [PubMed] [Google Scholar]
- 57.Suzuki K, Oguro H, Yamakawa T, Yamamoto A, Akao S, Saeki Y. 2008. Diversity and distribution of indigenous soybean-nodulating rhizobia in the Okinawa islands, Japan. Soil Sci Plant Nutr 54:237–246. 10.1111/j.1747-0765.2007.00236.x. [DOI] [Google Scholar]
- 58.Minami M, Yamakawa T, Yamamoto A, Akao S, Saeki Y. 2009. Estimation of nodulation tendency among Rj-genotype soybeans using the bradyrhizobial community isolated from an Andosol. Soil Sci Plant Nutri 55:65–72. 10.1111/j.1747-0765.2008.00333.x. [DOI] [Google Scholar]
- 59.Moawad HA, Ellis WR, Schmidt EL. 1984. Rhizosphere response as a factor in competition among three serogroups of indigenous Rhizobium japonicum for nodulation of field-grown soybeans. Appl Environ Microbiol 47:607–612. 10.1128/AEM.47.4.607-612.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Starkenburg SR. 2007. An investigation of carbon and nitrogen metabolism through a genomic analysis of the genus Nitrobacter. PhD Thesis. Oregon State University, Corvallis, OR. [Google Scholar]
- 61.De Meyer SE, Coorevits A, Willems A. 2012. Tardiphaga robiniae gen. nov., sp. nov., a new genus in the family Bradyrhizobiaceae isolated from Robinia psuedoacacia in Flanders (Belgium). Syst Appl Microbiol 35:205–214. 10.1016/j.syapm.2012.02.002. [DOI] [PubMed] [Google Scholar]
- 62.Ibiene AA, Agogbua JU, Okonko IO, Nwachi GN. 2012. Plant growth promoting rhizobacteria (PGPR) as biofertilizer: effect on growth of Lycopersicum esculentus. J Am Sci 8:318–324. [Google Scholar]
- 63.Safronova VI, Kuznetsova IG, Sazanova AL, Kimeklis AK, Belimov AA, Andronov EE, Pinaev AG, Pukhaev AR, Popov KP, Akopian JA, Willems A, Tikhonovich IA. 2015. Extra-slow-growing Tardiphaga strains isolated from nodules of Vavilovia formosa (Stev.). Arch Microbiol 197:889–898. 10.1007/s00203-015-1122-3. [DOI] [PubMed] [Google Scholar]
- 64.Streng A, Camp R, Bisseling T, Geurts R. 2011. Evolutionary origin of rhizobium Nod factor signaling. Plant Signal Behav 6:1510–1514. 10.4161/psb.6.10.17444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Marcondes de Souza JA, Alves LMC, Varani AM, Lemos EGM. 2014. The family Bradyrhizobiaceae, p 135–154. In Rosenberg E, DeLong EF, Lory S, Stackebrandt E, Thompson F (ed), The prokaryotes: prokaryotic biology and symbiotic associations, 4th ed, vol 1. Springer, Berlin, Germany. 10.1007/978-3-642-30197-1_253. [DOI] [Google Scholar]
- 66.Chizhevskaya EP, Naidenova EA, Onishchuk OP, Andronov EE, Simarov BV. 2018. The melanin biosynthesis gene from the CA15-1 strain of alfalfa nodule bacteria: molecular analysis and phylogeny. Russ J Genet 54:925–932. 10.1134/S1022795418080045. [DOI] [Google Scholar]
- 67.Dalal J, Kulkarni N. 2013. Antagonistic and plant growth promoting potentials of indigenous endophytic bacteria of soybean (Glycine max L. Merr). Curr Res Microbiol Biotechnol 1:62–69. [Google Scholar]
- 68.Vargas-Díaz AA, Ferrera-Cerrato R, Silva-Rojas HV, Alarcón AA. 2019. Isolation and evaluation of endophytic bacteria from root nodules of Glycine max L. (Merr.) and their potential use as biofertilizers. Span J Agric Res 17:e1103. 10.5424/sjar/2019173-14220. [DOI] [Google Scholar]
- 69.Monsanto Technology LLC. 2017. Seed applied solutions for soybean. Acceleron, Creve Coeur, MO. https://www.mssoy.org/uploads/files/acceleron-for-soybean.pdf [Google Scholar]
- 70.Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG, Van Horn DJ, Weber CF. 2009. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75:7537–7541. 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mirza BS, Sorensen DL, McGlinn DJ, Dupont RR, McLean JE. 2017. Dehalococcoides and general bacterial ecology of differentially trichloroethene dechlorinating flow-through columns. Appl Microbiol Biotechnol 101:4799–4813. 10.1007/s00253-017-8180-1. [DOI] [PubMed] [Google Scholar]
- 72.Wang Q, Garrity GM, Tiedje JM, Cole JR. 2007. Naïve Bayesian classifier for the rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 73:5261–5267. 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 35:1547–1549. 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mirza BS, Daniel J, McGlinn DJ, Brendan BJM, Nüsslein K, Tiedje JM, Rodrigues JLM. 2020. Diazotrophs show signs of restoration in Amazon rain forest soils with ecosystem rehabilitation. Appl Environ Microbiol 86:e00195-20. 10.1128/AEM.00195-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Oksanen J, Blanchet FG, Friendly M, Kindt R, Legendre P, McGlinn D. 2015. vegan: Community Ecology Package. R package version 2.4–1. http://CRAN.R-project.org/package=vegan.
- 76.Anderson MJ, Walsh DC. 2013. Permanova, Anosim, and the Mantel test in the face of heterogeneous dispersions: what null hypothesis are you testing? Ecol Monogr 83:557–574. 10.1890/12-2010.1. [DOI] [Google Scholar]
- 77.Brown JR (ed). 1998. Recommended chemical soil test procedures for the north central region. Missouri Agricultural Experiment Station SB 1001, University of Missouri, Columbia, MO. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The 16S rRNA gene sequences were deposited in the NCBI Sequence Read Archive (SRA) under BioProject ID PRJNA625756.