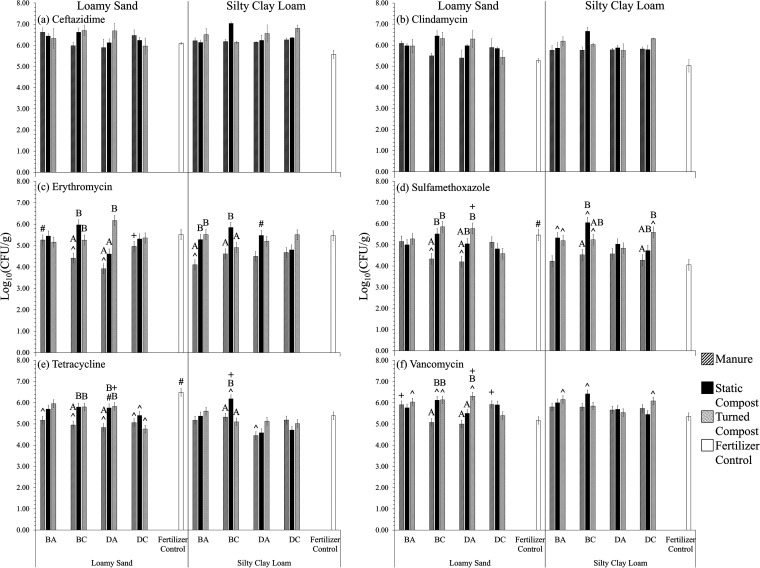
FIG 6.
Aerobic heterotrophic bacterial counts (HPCs) from radish taproot surfaces (n = 3) on R2A supplemented with (a) ceftazidime (25 μg/ml), (b) clindamycin (10 μg/ml), (c) erythromycin (25 μg/ml), (d) sulfamethoxazole (100 μg/ml), (e) tetracycline (3 μg/ml), and (f) vancomycin (11 μg/ml). Loamy sand (LS) and silty clay loam (SCL) were mixed with amendment from antibiotic-treated beef cattle (BA), antibiotic-free beef cattle (BC), antibiotic-treated dairy cattle (DA), or antibiotic-free dairy cattle (DC) or mixed with chemical fertilizer only as a control. Survival models were incorporated assuming a Weibull distribution to calculate the means and standard errors for Em, Smz, Tc, and Vm (indicated by the error bars). Standard errors for Cm and Cz are indicated by the error bars. Significant differences (P < 0.050, survival analysis) are indicated as follows: #, for the same amendment, the soil texture that yielded significantly greater gene abundance; ^, for the same soil texture, amendments that yielded gene abundances significantly different from that of the fertilizer control; +, for the same cattle type (beef or dairy), same amendment type, and same soil texture, the antibiotic condition (antibiotic or control) that yielded significantly greater gene abundance. The letters A and B indicate statistical groupings for effect of amendment type (manure, static compost, and turned compost) for the same cattle and same soil texture.