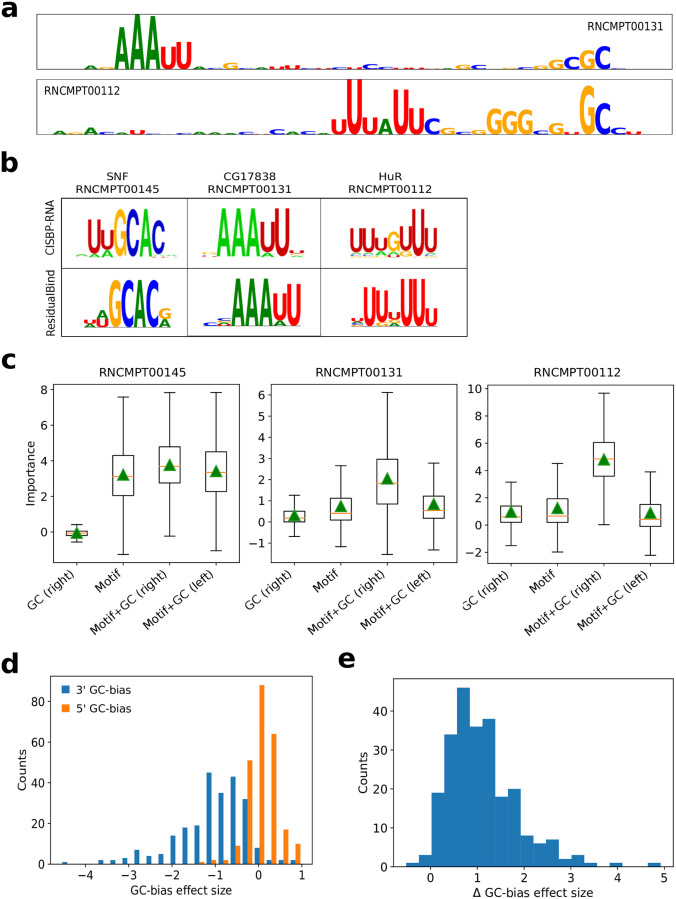
Fig 4. GC-bias in high binding score sequences.
(a) Representative sequence logos from in silico mutagenesis analysis for a test sequence with a top-10 binding score prediction for RNAcompete experiments for CG17838 (RNCMPT00131) and HuR (RNCPT00112). (b) Motif comparison between CISBP-RNA and ResidualBind’s motif representations generated by k-mer alignments. (c) Box plot of local importance for synthetic sequences with the top scoring 6-mer embedded in position 18-24 and GCGCGC embedded at positions 1-7 (Motif+GC, left) or positions 35-41 (Motif+GC, right). As a control, the GC content embedded at positions 35-41 without any motif is also shown. Green triangles represent the global importance. (d) Histogram of the GC-bias effect size, which is defined as the global importance when GC-bias is placed on the 5’ end (orange) and the 3’ end (blue) of synthetic sequences with a top scoring 6-mer embedded at positions 18-24 divided by the global importance of the motif at the center without any GC content, for each 2013-RNAcompete experiment. (e) Histogram of the difference between the GC-bias effect size, GC-bias on the 5’ end minus the 3’ end for each 2013-RNAcompete experiment.