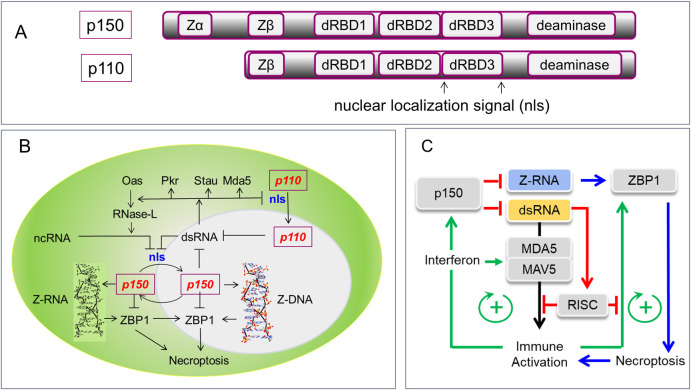
Fig 1. ADAR1 and Z-RNA.
(A) Both p150 and p110 ADAR1 isoforms contain the same deaminase domain and dRBDs. Only ADAR1 p150 contains the Zα domain that binds both Z-DNA and Z-RNA. The function of the Zβ domain is not known. (B) A cell with the cytoplasm in green and the nucleus shaded gray. While p110 is mostly nuclear, p150 undergoes cytoplasmic shuffling. Both isoforms have a bipartite nls that spans the third dsRBD3 B. The nls is inhibited by dsRNA (indicated by a “T” shaped arrow) that can be generated by nuclear export or from cytoplasmic nuclease processing of misfolded or noncoding RNAs. Cytoplasmic dsRNA can also affect other pathways by targeting MDA5 (activates interferon responses), Stau (Staufen decay pathway) [40], Pkr (effects on RNA translation) [41], and Oas (2′-5′-oligoadenylate synthetase that activates RNase-L to cleave noncoding RNAs like tRNA, Y-RNA, and ribosomal RNA) [42]. (C) MDA5 bound to dsRNA activates the MAVS protein to greatly increase the expression of interferon, ADAR1 p150, ZBP1, and other genes. ADAR1 p150 destabilizes dsRNA and inhibits ZBP1 induction of necroptosis [43]. The RISC uses miRNAs to target self-sequences. Both the formation of miRNAs and availability of target sequences are modulated by helicases [99,100]. dRBD, double-stranded RNA binding domain; dsRBD3, dsRNA binding domain 3; MAVS, mitochondrial antiviral signaling; MDA5, melanoma differentiation–associated gene 5; miRNA, microRNA; nls, nuclear localization signal; Pkr, protein kinase R; RISC, RNA-induced silencing complex; ZBP1, Z-DNA binding protein 1.