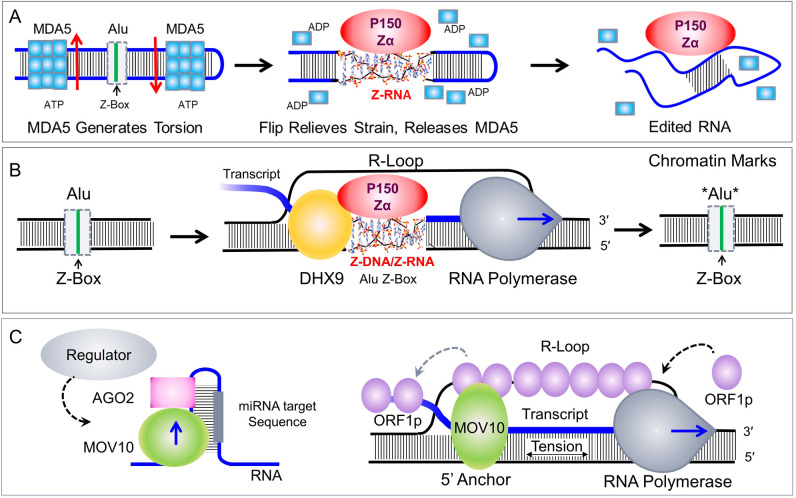
Fig 3. Different roles for helicases in self-recognition, with RNA strands drawn in blue and DNA strands in black.
(A) It is proposed that MDA5 induces Z-RNA formation in dsRNA by twisting and shortening the dsRNA. The strain induced promotes the flip to the longer Z-RNA helix, acting like a pull-switch to stop MDA5 filament formation. Alu retroelements contain a highly conserved Z-Box with sequences that favor Z-formation. Once Z-RNA is engaged by its Zα domain, ADAR1 deaminates adenosine to form inosine, destabilizing the dsRNA and preventing MDA5 filament formation. Both outcomes prevent the activation of interferon responses. (B) It is proposed that another helicase, DHX9, anchors R-loops when splicing is inhibited, localizing ADAR1 p150 to Alu Z-boxes in underwound regions of the DRH where Z-formation occurs, altering the transcript produced. Epigenetic marks on Alu chromatin following transcription are indicated by an asterisk—they threshold future responses. (C) The MOV10 normally unwinds dsRNA to allow the access of miRNAs and AGO2, a component of the RISC to their target sequences. This mechanism is one where specific self-sequences recognition enables suppression of immune responses in the normal state [100]. Alternatively, proteins, such as and FMRP [99] stabilize the MOV10 complex with its substrate and protects transcripts from suppression by RISC. It is proposed that a similar type of interaction with the LINE LI ORF1p locks MOV10 in place, anchoring the 5′ end of the transcript. The DRH formed are targets for RNaseH2. As described in the text, segments within the DRH may flip to the Z-conformation, causing ZBP1 activated necroptosis. With the helicase fixed in place, the movement of RNA polymerase tensions the element, powering the flip. AGO2, Argonaute2; DHX9, DExH-box helicase 9; DRH, DNA:RNA hybrid; dsRNA, double-stranded RNA; FMRP, fragile X mental retardation protein; MDA5, melanoma differentiation–associated gene 5; miRNA, microRNA; MOV10, Mov10 RISC complex RNA helicase; ORF1p, open reading frame 1 protein; RISC, RNA-induced silencing complex.