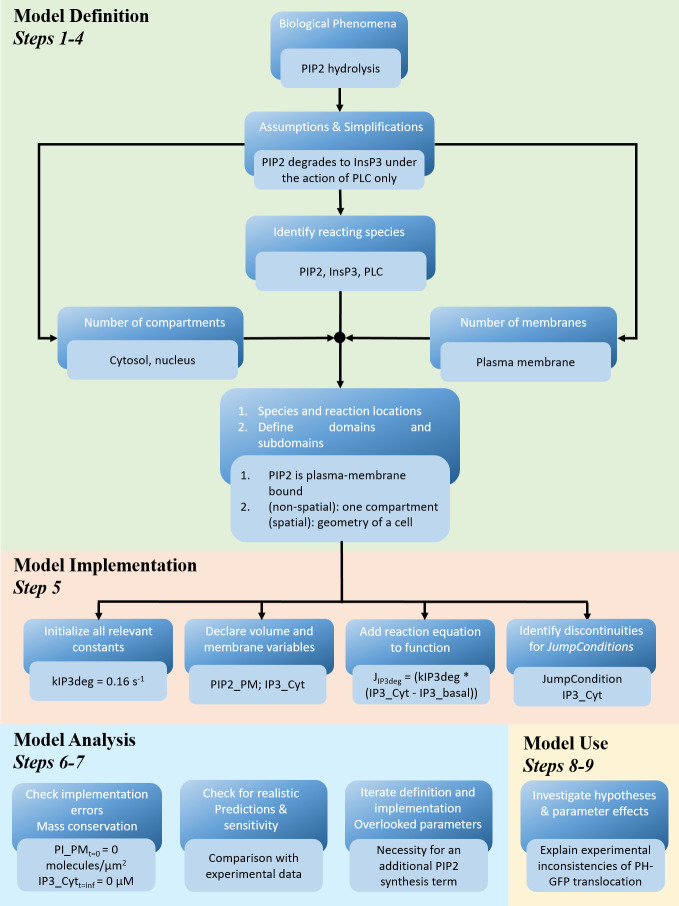
Fig 1. Schematic displaying the workflow for setting up a mathematical model.
Before starting the model implementation, modelers must define and simplify the biological phenomena to be simulated. The modeler sets the constants to be called in the equations, defines the membrane and compartment variables, adds needed mathematical expressions as functions, and finally defines all the fluxes at membranes as jump conditions. After the model implementation, the model can be checked for implementation errors and validated, and then used to investigate hypotheses in silico. The dark blue boxes represent the generic workflow, whereas the light blue boxes apply these to the phosphoinositide turnover kinetics example [14].