Abstract
Pseudohypoparathyroidism (PHP) and pseudopseudohypoparathyroidism (PPHP) are caused by mutations and/or epigenetic changes at the complex GNAS locus on chromosome 20q13.3 that undergoes parent-specific methylation changes at several differentially methylated regions (DMRs). GNAS encodes the alpha-subunit of the stimulatory G protein (Gsα) and several splice variants thereof. PHP type Ia (PHP1A) is caused by heterozygous inactivating mutations involving the maternal exons 1-13. Heterozygosity of these maternal GNAS mutations cause PTH-resistant hypocalcemia and hyperphosphatemia because paternal Gsα expression is suppressed in certain organs thus leading to little or no Gsα protein in the proximal renal tubules and other tissues. Besides biochemical abnormalities, PHP1A patients show developmental abnormalities, referred to as Albright’s hereditary osteodystrophy (AHO). Some, but not all of these AHO features are encountered also in patients affected by PPHP, who carry paternal Gsα-specific mutations and typically show no laboratory abnormalities. Autosomal dominant PHP type Ib (AD-PHP1B) is caused by heterozygous maternal deletions within GNAS or STX16, which are associated with loss of methylation at the A/B DMR alone or at all maternally methylated GNAS exons. Loss of methylation of exon A/B and the resulting biallelic expression of A/B transcript reduces Gsα expression thus leading to hormonal resistance. Epigenetic changes at all differentially methylated GNAS regions are also observed in sporadic PHP1B, which is the most frequent PHP1B variant. However, this disease variant remains unresolved at the molecular level, except for rare cases with paternal uniparental isodisomy or heterodisomy of chromosome 20q (patUPD20q).
Keywords: pseudohypoparathyroidism, PTH, calcium, phosphate, cAMP, TSH, Gs-alpha, GNAS, STX16, epigenetics, parent-specific GNAS methylation
This review will focus on pseudohypoparathyroidism type Ia (PHP1A) and the different variants of pseudohypoparathyroidism type Ib (PHP1B) with some discussion of pseudopseudohypoparathyroidism (PPHP) and its variant progressive osseous heteroplasia (POH), namely disorders that are all caused by impaired agonist-dependent 3′,5′-cyclic adenosine monophosphate (cAMP) formation (1-5). The genetic defects that cause different forms of acrodysostosis will not be discussed, although some are caused by mutations in genes such as PTHLH, PRKAR1A, PDE4D, PDE3A, or HDAC4, which impair cAMP/protein kinase A signaling (6-11). Consequently, patients affected by acrodysostosis can have clinical, radiographic and/or laboratory features reminiscent of PHP1A, PHP1B, or PPHP. The first genetic defects responsible for PHP1A and PPHP, namely heterozygous mutations involving the GNAS exons encoding the alpha-subunit of the stimulatory G protein (Gsα), were discovered more than 30 years ago. Much later the first genetic causes of PHP1B were identified, but most variants remain undefined at the molecular level. In fact, besides a rare form of sporadic PHP1B (sporPHP1B), only autosomal dominant PHP1B (AD-PHP1B) subtypes have been defined at the genetic and epigenetic level. SporPHP1B is the most frequent form of the disorder and although GNAS methylation changes are routinely assessed to establish the diagnosis of this PHP1B variant, the underlying genetic defects remain to be defined for most cases. In conjunction with the biochemical and epigenetic characterization of several genetically altered mouse strains, the molecular defects and GNAS methylation changes leading to the different PHP1B variants have enhanced the understanding of the underlying disease mechanisms that most likely involve abnormalities during oocyte development. These insights from mice with distinct mutations and from patients affected by PHP1B will also have implications for explaining the different abnormalities encountered in PHP1A and PPHP. This review will summarize the known genetic and epigenetic defects that cause the different forms of PHP and the insights gained into the intricate regulation at the GNAS locus that encodes Gsα and several additional transcripts.
First Description of a Novel Syndrome
The term pseudohypoparathyroidism (PHP) was first introduced in 1942 by Albright et al. to describe several patients who presented with hypocalcemia and hyperphosphatemia in association with obesity, short stature, short metacarpals and metatarsals, as well as neurocognitive impairment, namely clinical findings that are now referred to as Albright’s hereditary osteodystrophy (AHO) (12). The affected individuals showed no phosphaturic response to parathyroid extracts and the authors therefore concluded that the abnormal regulation of mineral ion homeostasis was due to renal resistance to parathyroid hormone (PTH) rather than PTH deficiency. These first descriptions of PHP patients laid the ground work for defining over the next several decades a more complete spectrum of the clinical and laboratory abnormalities associated with this rare and variable disorder.
First Insights: Impaired PTH-stimulated cAMP Generation in Pseudohypoparathyroidism
Shortly after the discovery that epinephrin and glucagon stimulate the generation of cAMP in liver homogenates (13), Aurbach and colleagues revealed that PTH increases the formation of this second messenger in plasma membranes from kidney and bone tissue (14-16). These authors had also shown that injection of PTH preparations into animals increases the urinary excretion of cAMP, and, with some delay, the urinary excretion of phosphate (17). They subsequently determined that PTH treatment rapidly increases urinary cAMP excretion in healthy humans and in patients with postsurgical hypoparathyroidism, but not in patients affected by PHP (18). These studies suggested a partial or total lack of PTH-sensitive adenyl cyclase in the kidney of PHP patients and furthermore linked lack of the renal PTH-stimulated cAMP response to the blunted phosphaturic response noticed earlier by Albright et al. (12).
G-protein Deficiency Due to Heterozygous Gsα Mutations Causes PHP
In 1980, Farfel et al. (19) as well as Levine et al. (20) investigated extracts of erythrocyte membranes from patients who are now known to be affected by PHP1A, namely patients with characteristic AHO features, hypocalcemia, and impaired urinary excretion of cAMP and phosphate in response to exogenous PTH. Both groups of investigators revealed an approximately 50% reduction in the activity of a guanine nucleotide-binding protein (G-protein), later shown to be Gsα that couples various G protein–coupled receptors (GPCRs) to adenylate cyclase and hence the formation of cAMP (1, 2). Because of the combination of PTH-resistant hypocalcemia and hyperphosphatemia, impaired PTH-stimulated urinary cAMP and phosphate excretion, AHO features, and reduced Gsα activity, this disorder is now referred to as PHP1A. Surprisingly, cell membranes from patients affected by PPHP (21) exhibited a similar reduction in G-protein activity as cells from patients affected by PHP1A, despite normal calcium and phosphate levels (see below).
After the molecular cloning of cDNAs encoding human Gsα (22) and defining the structure of its gene (GNAS) (23), nucleotide sequence analyses of genomic DNA from several PHP1A and PPHP patients revealed the first disease-causing mutations involving those GNAS exons that encode Gsα (24, 25). These GNAS mutations are heterozygous thus explaining, for both groups of patients, the approximately 50% reduction of Gsα protein and activity, which was observed in readily accessible cells of the patients but is expected to occur in all tissues (1, 2). GNAS mutations had provided essential first insights into the genetic defects underlying PHP1A. However, it remained uncertain as to why heterozygous Gsα mutations should lead to disease at all, until Davies and Hughes (26) documented that GNAS mutations involving exons 1-13 lead to AHO in combination with PTH-resistant hypocalcemia if inherited from a female, while inheritance of such mutations from a male leads only to AHO features, but not to hormonal resistance, that is PPHP. By now numerous Gsα mutations have been identified as causes of the different PHP variants (27); these are located on the maternal GNAS allele in patients affected by PHP1A and on the paternal GNAS allele in patients affected by PPHP (for databases listing the known mutations see www.hgmd.cf.ac.uk/ac/all.php or www.lovd.nl/GNAS).
A PHP Variant Without Apparent G-protein Deficiency
Besides reduced G-protein activity in PHP1A, Farfel et al. (19) reported normal activity levels for red blood cell membranes obtained from patients affected by postsurgical hypoparathyroidism, who showed cAMP and phosphate excretion in response to PTH, and from patients affected by PHP type II (PHP2), who showed, in response to PTH, an increase in urinary cAMP, but not in urinary phosphate excretion (28). Importantly, the authors also reported findings in a PHP variant in which patients present with hypocalcemia, elevated PTH levels, and impaired excretion of cAMP and phosphate in response to PTH, yet no AHO features. Samples from those patients exhibited normal G-protein activity, and therefore, this PHP variant has been referred to as PHP type Ib (PHP1B) (2).
The GNAS Complex Locus Gives Rise to Different Variants of Gsα and to Several Additional Transcripts
GNAS is a complex genetic locus that gives rise to several different transcripts (3) (Table 1). The best known and most widely studied GNAS-encoded protein is Gsα, which comes in 4 different variants that are derived from the same primary transcript initiated at exon 1. The long version of Gsα is encoded by exons 1-13, while the short version lacks the portion encoded by exon 3 and additional complexity is generated by the presence or absence of an extra CAG codon between the nucleotide sequences derived from exons E3 and E4 (22, 23). These transcripts were shown to be expressed at different levels in some tissues and although there is little evidence for functional differences between the 4 Gsα proteins (29, 30), a mutation in exon 3 that truncates the long, but not the short version of Gsα, causes PHP1A (31).
Table 1.
Alternative first GNAS exons giving rise to different RNA transcripts and proteins
| Normal methylation | Autosomal dominant PHP1B caused by maternal mutations | Sporadic PHP1B | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Deletion of portions of STX16 or of exon NESP | Deletion of exons AS3-4 with or without NESP | Genetic defect unknown | ||||||||
| Exons with promoters | RNA transcripts | Proteins | Mat | Pat | Mat | Pat | Mat | Pat | Mat | Pat |
| NESP | NESP* → exons 2-13 | NESP55 | – | + | – or 0 | + | – or 0 | + | + | + |
| AS | AS exon 1-5 | non-coding | + | – | + | – | – | – | (–) | – |
| XL | XL → exons 2-13 | XLαs | + | – | + | – | – | – | (–) | – |
| A/B | A/B → exons 2-13 | N-truncated Gsα | + | – | – | – | – | – | – | – |
| Gsα-exon 1 | exons 1-13 (±exon 3; ±CAG) | Gsα-long/short ± Gln | Not methylated | Reduced or no Gsα expression from the paternal GNAS allele in some tissues |
With exception of the unmethylated Gsα exon 1, the promoters of the alternative first exons undergo parent-specific methylation in healthy individuals. Maternal deletions involving STX16 or NESP are the cause of some autosomal dominant PHP1B variants that are associated with loss of methylation at exon A/B, while maternal deletions that comprise exons AS3-4 with or without deletion of NESP lead to loss of all 3 maternal GNAS methylation imprints. In sporadic PHP1B there is loss of A/B methylation, often variable loss of methylation at the AS and XL, and gain of methylation at NESP. Some tissues, such as the proximal renal tubules, show reduced or no Gsα expression from the paternal GNAS allele, but the underlying silencing mechanism remains to be determined. *, termination codon in exon NESP.
Abbreviations: AS, antisense transcript; Gsα, alpha-subunit of the stimulatory G protein; PHP1B, pseudohypoparathyroidism type Ib
Besides the 4 Gsα variants, 3 additional mRNAs are derived from distinct first exons and their promoters that splice onto GNAS exons 2-13. These alternative transcripts are A/B, extra-large form of Gsα (XLαs), and neuroendocrine secretory protein 55 (NESP55). The A/B transcript was initially thought to be nontranslated because exons A/B and 2 comprise no initiator methionine. However, through the use of an ATG in exon 3, the A/B transcript was subsequently shown to give rise to an amino-terminally truncated Gsα that reduces activity of the full-length G protein (32). The XLαs transcript encodes an extra-large Gsα variant comprising a unique protein sequence derived from exon XL followed by the portion of Gsα that is encoded by exons 2-13 (33). The NESP55 transcript encodes a neuroendocrine secretory protein with a molecular weight of 55 kDa; because of a termination codon within the first exon for this transcript, the nucleotide sequence derived from GNAS exons 2-13 are part of the 3′-noncoding region (34). In addition to these alternatively spliced sense mRNAs, an antisense transcript (AS) is derived from a fifth promoter on the opposite DNA strand (35). Furthermore, use of an alternative transcriptional start site within the XL exon can lead to a distinct mRNA and a protein termed XXLαs. Moreover, both XLαs and XXLαs transcripts contain a second open frame leading to gene products named ALEX or ALEXX. The ALEX protein has been shown to interact directly with XLαs (36).
Maternal Gsα Expression is Required for Normal Hormonal Responsiveness in Several Tissues
In 1969, as outlined above, Chase and colleagues showed that PPHP patients treated with PTH have a robust increase in urinary cAMP excretion that was indistinguishable from that in healthy controls and in patients affected by postsurgical hypoparathyroidism (18). Such patients were later shown to be carriers of heterozygous, loss-of-function Gsα mutations on the paternal GNAS allele, while patients affected by PHP1A carry such mutations on their maternal GNAS allele (Fig. 1). Heterozygosity for these mutations thus explained the equivalent reduction in G-protein activity observed for plasma membranes prepared from red blood cells or skin fibroblasts of patients affected by either PHP1A or PPHP. Importantly, these data indicated that Gsα expression from the maternal GNAS allele is essential for mediating PTH-induced urinary cAMP excretion, which is stimulated by the actions of PTH in the proximal renal tubules. These findings are consistent with the conclusion that this portion of the kidney relies on Gsα derived from the maternal GNAS allele. Subsequently, it was furthermore shown that Gsα is derived predominantly from the maternal allele in thyroid, gonads, pituitary, portions of the central nervous system, and brown adipose tissue. In these tissues, paternal Gsα expression is silenced through an as yet undefined mechanisms (37-43).
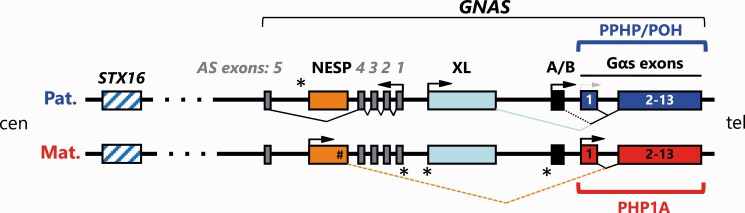
Figure 1.
Organization of the GNAS locus. The GNAS complex gives rise to several imprinted sense and antisense transcripts. Gsα, the most abundant product derived from this locus, is encoded by exons 1-13. This transcript is mostly biallelically expressed, except for few tissues like renal proximal tubule, thyroid, pituitary, brown adipose tissue, gonads, and various nuclei in the brain where paternal Gsα expression is partially or completely silenced through yet undefined mechanisms. The GNAS locus gives rise to 4 other transcripts, including the antisense transcript (AS), the NESP transcript encoding the neuroendocrine secretory protein 55 (NESP55), as well as the transcript encoding an extra-large form of Gsα, named XLαs, and the A/B transcript (1A in mice) that may give rise to an amino-terminally truncated Gsα. Promoters for XL, A/B and AS transcripts are methylated on the maternal allele and thus transcribed exclusively from the paternal allele. The NESP promoter is paternally methylated and active exclusively on the maternal allele. The NESP, XLαs, and A/B transcripts are derived from unique first exons that splice onto GNAS exon 2-13. Immediately centromeric of the XL promoter lies the promoter for the antisense transcript AS. Mutations involving any of the thirteen GNAS exons encoding Gsα are associated with AHO (Albright’ hereditary osteodystrophy) with or without hormone resistance based on which parental allele is affected, namely pseudohypoparathyroidism 1A (PHP1A), if a mutation is present on the maternal allele (horizontal red bracket), or pseudopseudohypoparathyroidism (PPHP) and progressive osseus heteroplasia (POH), if a mutation is present on the paternal allele (horizontal blue bracket). Maternal and paternal GNAS-derived mRNAs are shown above and below the gene structure. Boxes indicate exons and splicing patterns are represented by broken/angled lines. Bent arrows indicate promoter start sites and direction of transcription. *Sites of differentially methylated regions (DMRs) on the maternal (XL, A/B, and AS) and the paternal (NESP) promoters.
Silencing of paternal Gsα expression develops gradually, at least in the proximal renal tubules. Illustrating this, a PHP1A patient with a maternal GNAS mutation was shown to have a normal increase in PTH-stimulated urinary cAMP excretion at the age of 7 months, but then a markedly blunted response at the age of 3.9 years (44). Similarly, a patient with PHP1B revealed normal plasma PTH concentrations until the age of 2 years, when levels were shown to be elevated as evidence for acquired resistance to the hormone (45). Furthermore, studies in mice with maternal deletion of Gnas exon 1 revealed that paternal Gsα protein/mRNA expression in the proximal renal tubules declines over the first 2 months of life thus resulting gradually in PTH resistance and increased circulating PTH levels (46). Taken together, these findings indicate that Gsα expression in the renal proximal tubular cells is biallelic early in life, but the paternal contribution to total Gsα protein levels subsequently declines thereby explaining the delayed development of PTH resistance in the absence of maternally derived Gsα.
GNAS Undergoes Parent-specific Epigenetic Changes at Several Differentially Methylated Regions
GNAS is one of only few genetic loci that undergo parent-specific changes in DNA methylation to limit protein expression to only one parental allele (33, 35, 47). The DMRs involving the promoters of exons AS, XL, and A/B are methylated only on the maternal GNAS allele; consequently, transcription from these promoters occurs only from the non-methylated paternal allele. The NESP promoter is methylated on the paternal allele and thus its mRNA is transcribed only maternally. In contrast, the Gsα promoter at exon 1 does not undergo parent-specific methylation. Nonetheless, its transcription is not always biallelic.
Human GNAS is located on the long arm of chromosome 20, while mouse Gnas is located on chromosome 2 (23, 48). In both species, the 4 DMRs undergo indistinguishable methylation changes and the corresponding promoters give rise to the same series of sense and antisense transcripts (33, 35, 49). Conclusions based on findings in mice with genetic alterations in Gnas, and in humans affected by PHP variants that are caused by different GNAS mutations have provided complementary insights into the underlying mechanisms by which Gsα expression is regulated in normal health and dysregulated in disease.
Pseudohypoparathyroidism Type Ia
PTH-resistant hypocalcemia and hyperphosphatemia are the most prominent features of PHP1A, but patients affected by this disorder can also develop resistance towards several additional hormones, including thyrotropin, calcitonin, and growth hormone–releasing hormone (2, 50, 51) and possibly melanocortin (52). Besides the AHO features, several additional clinical abnormalities are likely to be associated with a lack of maternal Gsα, including hearing loss, decreased olfaction, sleep apnea, and asthma-like symptoms, thus implicating abnormal signaling at several additional GPCRs (4, 53). While several reports had provided no evidence for an obvious relationship between genotype and phenotype for PHP1A patients (27, 54-56), recent findings have indicated that PHP1A patients with deleterious truncating mutations show significantly more subcutaneous calcifications than patients with missense mutations. In contrast, foreshortened metacarpals/-tarsals were more frequently encountered with missense mutations, while obesity, short stature, and neurocognitive defects did not vary for patients with the different kinds of GNAS mutations (57).
Because of the parent-specificity of GNAS mutations and the involvement of different agonists acting through various distinct GPCRs, it is likely that a recently proposed new nomenclature for PHP and related diseases needs to be revised further because it focuses primarily on defects caused by impaired PTH- and PTHrP-dependent signaling, and does not take into account resistance towards other agonists and the parental allele carrying the GNAS mutation (58). However, the outlined guidelines have shown considerable promise by relying on few major clinical signs to suspect the diagnosis before proceeding with additional clinical and genetic assessment (59). Furthermore, the previously established nomenclature has equally severe limitations as the new one as it implies that endocrine abnormalities are restricted to PTH resistance and the term osteodystrophy refers to multiple abnormalities that affect not only the skeleton, but also neurocognition, weight regulation, and other developmental defects. Thus a further modified classification should be developed that focuses on impaired cAMP signaling rather than resistance to individual hormones.
Pseudopseudohypoparathyroidism
PPHP was also described first by Albright et al., when this group of investigators presented a patient with typical AHO features, yet without evidence for hormonal resistance and thus normal levels of serum calcium and phosphate (21). PPHP patients are now known to have some AHO features, such as short metacarpals and metatarsals, short stature, and round face, but usually without neurocognitive abnormalities and without obesity (4, 60-62), and only rare cases present with evidence for mild hormonal resistance (63). Furthermore, at birth individuals who are later diagnosed with PPHP are typically small for gestational age, particularly when paternal GNAS mutations affect exons 2-13, which suggests that paternal XLαs may have an important role during fetal development (64, 65). This conclusion is supported by findings in mice with targeted ablation of the XL exon, as these animals exhibit poor postnatal growth and survival (66). In addition, patients with maternal uniparental disomy of chromosome 20q (matUPD20q), who lack XLαs expression because the 2 maternal XL DMRs are both methylated, present with pre- and postnatal growth failure, hyperactivity, severe feeding difficulties, and short stature (67-69).
In addition to intrauterine and postnatal growth retardation and the subsequent development of several AHO features, some patients with paternal GNAS mutations develop POH. This more severe variant of PPHP is frequently caused by mutations involving the paternal GNAS exons 2-13, less often by exon 1 mutations. This also raises the question whether POH is related primarily to deficiency of paternal XLαs protein, rather than deficiency of paternal Gsα (5, 27, 70-72).
Different variants of pseudohypoparathyroidism type Ib
PHP1B occurs either as an autosomal dominant disorder or as a sporadic disease (AD-PHP1B and sporPHP1B, respectively). As in PHP1A, PTH resistance in renal proximal tubules and thus hypocalcemia and hyperphosphatemia despite elevated plasma PTH levels are the most obvious laboratory abnormalities in PHP1B. However, some resistance to thyrotropin is frequently noticed (4, 40, 73-76). PHP1B patients can also present with mild AHO features (4, 77-80) or even show bone abnormalities that are indistinguishable from those encountered in PHP1A or PPHP (81, 82). Analyses of erythrocyte membranes from PHP1B patients typically reveal normal Gsα bioactivity, which is different from the findings in PHP1A and PPHP; however, few PHP1B patients, particularly those with AHO features, show reduced Gsα activity, indicating that a threshold for Gsα activity could be required for the development of some of these clinical aspects (83).
Patients affected by PHP1B display accelerated fetal growth resulting at birth in significantly higher weights and increased lengths, while patients affected by PHP1A are slightly smaller than average (62, 84). The increase in birth weight is particularly pronounced for patients with AD-PHP1B due to the 3-kb STX16 deletion that results in loss of methylation at the exon A/B DMR, if these children are born to healthy female carriers of the genetic defect. Their birth weights differ from those of PPHP/POH patients with mutations involving GNAS exons E2-13 (65) by almost 4.5 standard deviation scores. This difference in birth parameters suggests a significant growth advantage due to loss of methylation at the exon A/B DMR (84).
Autosomal dominant forms of pseudohypoparathyroidism
A family in which several members had hypocalcemia and/or elevated PTH levels, but without obvious skeletal and other abnormalities, had been described by Winter and Hughes (85). Furthermore, in the report by Farfel et al. several members of a single family had presented with PTH-resistant hypocalcemia, but without AHO features and with normal G-protein activity in extracts from red blood cell membranes (19). In both kindreds, the disease followed an autosomal dominant mode of inheritance. Using genomic DNA from several subsequently identified families with autosomal dominant PHP1B (AD-PHP1B), the genetic defect was mapped to a region on chromosome 20q13.3 that is located centromeric of GNAS (86, 87). This led to the discovery of a recurrent 3-kb deletion in STX16, the gene encoding syntaxin 16, which is the most frequent cause of this PHP1B variant (3, 4, 88). Additional genetic mutations responsible for AD-PHP1B have since been identified, including several other deletions as well as different duplications and an inversion; these disease-causing mutations are all located within a region on the long arm of chromosome 20 that extends from STX16 to GNAS (Fig. 2).
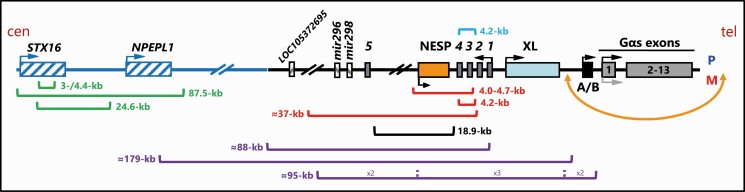
Figure 2.
Genetic causes of autosomal dominant pseudohypoparathyroidism type Ib (AD-PHP1B). AD-PHP1B due to maternal deletions upstream of the Gsα-coding region that are associated with loss of methylation at one or more DMRs. Maternal deletions of STX16 or NESP (green or black brackets) lead to loss of methylation specifically at the A/B DMR. The same epigenetic GNAS changes can also be due to a large genomic inversion (orange) or due to different duplications (purple brackets) involving the maternal allele. Deletion of AS exons 3-4 on the paternal allele (light blue bracket) leads to a partial loss of methylation at the paternal NESP DMR and a partial gain of methylation at the paternal A/B DMR. Maternal deletions that include AS exons 3-4 alone or in combination with exon NESP (red brackets) result in loss of methylation at all 3 maternal GNAS DMRs (A/B, XL, and AS). Boxes and connecting lines represent exons and introns, respectively. Arrows indicate direction of transcription. Brackets indicate deletions or duplications.
The 3-kb deletion as well as other STX16 deletions, if located on the maternal allele, are associated with loss of methylation at the A/B DMR, but not at the AS and XL DMRs (45, 89, 90). Interestingly, deletion of the equivalent deletion in mice does not lead to loss of methylation at Gnas (91), which raises the possibility that the cis-acting elements regulating methylation at A/B and at the corresponding mouse 1A DMR are spatially distinct in humans and mice. Methylation changes restricted to the A/B DMR were furthermore documented for a maternal deletion extending from the NESP exon and its promoter to a region telomeric of AS exon 5 (92). This 18.9-kb deletion completely eliminates the NESP exon suggesting that the transcript/protein derived from this promoter is required for establishing methylation at the exon A/B DMR (see below). Of note, the AS exons are not affected by this deletion, which is consistent with normal methylation at the AS and XL DMRs. Loss of methylation restricted to the A/B DMR can be caused also by different genetic duplications within GNAS (93-95). In addition, a large maternal inversion with the centromeric breakpoint located between exons XL and A/B leads to loss of methylation that is restricted to the A/B DMR (96).
In contrast to the STX16 deletions that cause methylation changes restricted to the A/B DMR, maternal deletions involving either exons AS3-4 alone, or exons NESP and AS3-4 (97-99) lead to loss of methylation at all 3 maternal DMRs, namely AS, XL, and A/B. Identical epigenetic changes are observed when a similar deletion is introduced into the mouse (100). Interestingly, deletion of exons AS3-4 leads, when located on the paternal allele, to a partial loss of methylation at NESP and an incomplete gain of methylation at exon A/B (99), thereby providing insights into the underlying mechanisms responsible for establishing GNAS methylation imprints (see below).
Sporadic PHP1B
Sporadic (spor)PHP1B patients present with laboratory abnormalities that are largely indistinguishable from those observed in AD-PHP1B (73, 98, 101-105). These cases show a loss of methylation at the maternal GNAS exons AS, XL, and A/B, and, importantly, all sporPHP1B patients show a gain of methylation at the NESP DMR. In numerous sporadic PHP1B cases, the methylation changes within GNAS are incomplete, which is different from the uniform epigenetic changes encountered in the familial forms of the disease (73, 78, 98, 106, 107). Unlike autosomal dominant forms of PHP1B, the molecular causes of most sporPHP1B patients have yet to be identified, except for those patients who have uniparental disomy involving the long arm of chromosome 20 (patUPD20q) (108-112).
Hypotheses Regarding the Mechanisms Leading to Abnormal GNAS Methylation
The GNAS methylation abnormalities that lead to the different familial and sporadic PHP1B variants occur most likely during oogenesis. Studies in mice show that the genome of oogonia undergoes complete demethylation during early oogenesis and subsequently 3 sites, namely AS, Xl, and 1A, are remethylated before meiosis I (113). The epigenetic defects that underly the different inherited PHP1B variants characterized by loss of methylation at one or more of the maternal GNAS DMRs, namely AS, XL, and A/B, can thus be explained by disruption of the re-methylation events that occur during oocyte development.
The mouse Nesp DMR, which is completely unmethylated in sperm, does not get re-methylated until embryonic day 10.5 (114). It is therefore likely that methylation of the paternal and the maternal NESP DMRs, as encountered in most sporPHP1B patients (73, 98, 101-105), occurs postfertilization. One PHP1B family has been described in which 2 affected sisters show GNAS methylation changes that are similar to those encountered in sporPHP1B, including a gain of methylation at the maternal NESP DMR. Interestingly, however, linkage to the STX16/GNAS region was excluded in this family (103). This makes it likely that the primary genetic defect predicted for the affected members of this kindred, and possibly in at least some sporPHP1B cases, occurs outside of GNAS elsewhere in the genome.
The definition of several PHP1B cases that are associated with STX16 or GNAS deletions, as well as findings in genetically modified mice provided important clues regarding the regulation of GNAS methylation. For example, a lack of paternal AS transcription caused by the introduction of a polyadenylation signal into AS exon 1 results in a lack of methylation at the paternal Nesp DMR and a concomitant gain of methylation at the paternal exon 1A (115, 116). Likewise, in one AD-PHP1B kindred, the deletion of exons AS3-4 on the paternal GNAS allele is associated with reduced methylation of the paternal NESP DMR and increased paternal A/B methylation. It therefore appears likely that a lack of full-length AS transcripts leads to active NESP transcription and hence methylation of the paternal A/B DMR (99). Taken together, findings in genetically altered mice and in one AD-PHP1B family show that active AS transcription is essential for enabling methylation at the NESP DMR.
In addition, a truncated maternal Nesp transcript in mice is associated with loss of methylation at the maternal 1A DMR (115, 116). Likewise, deletion of exon NESP on the maternal allele and thus a complete absence of NESP transcription, is associated in the affected members of an AD-PHP1B kindred with loss of methylation at exon A/B (92). These findings indicate that a maternal sense transcript derived from the NESP promoter is required for establishing methylation at the A/B DMR during oogenesis.
The importance of the GNAS locus for normal oocyte development is underlined by several recent findings. For example, Gsα was shown to play an essential role during oocyte maturation since Cre-mediated ablation of Gnas exon 1 under the control of the oocyte-specific Zp3 promoter causes complete infertility (117). Furthermore, females affected by PHP1A or PPHP (118) preferentially transmit the mutant GNAS allele to the next generation. Similar transmission ratio distortion is observed for females who are carriers of STX16 or GNAS mutations that cause AD-PHP1B when located on the maternal allele (119). In addition, females affected by AD-PHP1B due to a maternal mutation have significantly fewer offspring than unaffected females, who carry these mutations on their paternal allele. It is, however, unclear whether premature resumption of meiosis due to impaired Gsα expression is responsible for these latter findings.
In conclusion, the GNAS locus on chromosome 20q13.3, which encodes Gsα and several different splice variants thereof, is subject to complex regulatory mechanisms that include parent-specific methylation of several exons/promoters and a tissue-/cell-specific reduction in paternal Gsα expression through as-yet incompletely understood mechanisms. Three distinct, yet related diseases are caused by mutations in this genetic locus. PHP1A is caused by maternal mutations that directly impact the encoded Gsα protein, while PPHP results when the same or similar mutations are located on the paternal allele. The autosomal dominant forms of AD-PHP1B are caused by maternal deletions, duplications, or an inversion involving GNAS or STX16, which lead to distinct methylation changes involving one or several different GNAS DMRs. With the exception of patients with patUPD20q, most sporadic PHP1B cases are unresolved.
Acknowledgments
Many thanks to Drs. Thomas Gardella and Murat Bastepe for their careful review of the manuscript and their very helpful suggestions.
Financial Support: This work was supported by grants from the National Institutes of Health, NIDDK (DK046718 and DK11794, subproject III).
Glossary
Abbreviations
- cAMP
3′,5′-cyclic adenosine monophosphate
- AD-PHP1B
autosomal dominant PHP type Ib
- AHO
Albright’s hereditary osteodystrophy
- AS
antisense transcript
- DMR
differentially methylated region
- Gsα
alpha-subunit of the stimulatory G protein
- GPCR
G protein–coupled receptor
- PHP
pseudohypoparathyroidism
- PHP1A
PHP type Ia
- PHP1B
pseudohypoparathyroidism type Ib
- PHP2
pseudohypoparathyroidism type II
- POH
progressive osseous heteroplasia
- PPHP
pseudopseudohypoparathyroidism
- PTH
parathyroid hormone
Data Availability
Data sharing is not applicable to this article as no datasets were generated or analyzed during the current study.
References
- 1. Weinstein LS, Liu J, Sakamoto A, Xie T, Chen M. Minireview: GNAS: normal and abnormal functions. Endocrinology. 2004;145(12):5459-5464. [DOI] [PubMed] [Google Scholar]
- 2. Levine M. Hypoparathyroidism and pseudohypoparathyroidism. In: DeGroot LJ, Jameson LJ. ed. Endocrinology. 5th ed. W.B. Saunders Co; 2005. [Google Scholar]
- 3. Bastepe M, Jüppner H. Pseudohypoparathyroidism, Albright’s hereditary osteodystrophy, and progressive osseous heteroplasia: disorders caused by inactivating GNAS mutations. In: DeGroot LJ, Jameson JL, eds. Endocrinology. Vol. 1, 7th ed. W.B. Saunders Company; 2016:1147-1159. [Google Scholar]
- 4. Mantovani G, Bastepe M, Monk D, et al. Diagnosis and management of pseudohypoparathyroidism and related disorders: first international Consensus Statement. Nat Rev Endocrinol. 2018;14(8):476-500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Shore E, Ahn J, Jan de Beur S, et al. Paternally-inherited inactivating mutations of the GNAS1 gene in progressive osseous heteroplasia. New Engl J Med. 2002;346(2):99-106. [DOI] [PubMed] [Google Scholar]
- 6. Lee H, Graham JM Jr, Rimoin DL, et al. Exome sequencing identifies PDE4D mutations in acrodysostosis. Am J Hum Genet. 2012;90(4):746-751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Michot C, Le Goff C, Goldenberg A, et al. Exome sequencing identifies PDE4D mutations as another cause of acrodysostosis. Am J Hum Genet. 2012;90(4):740-745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Maass PG, Aydin A, Luft FC, et al. PDE3A mutations cause autosomal dominant hypertension with brachydactyly. Nat Genet. 2015;47(6):647-653. [DOI] [PubMed] [Google Scholar]
- 9. Klopocki E, Hennig BP, Dathe K, et al. Deletion and point mutations of PTHLH cause brachydactyly type E. Am J Hum Genet. 2010;86(3):434-439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Williams SR, Aldred MA, Der Kaloustian VM, et al. Haploinsufficiency of HDAC4 causes brachydactyly mental retardation syndrome, with brachydactyly type E, developmental delays, and behavioral problems. Am J Hum Genet. 2010;87(2):219-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Reyes M, Bravenboer B, Jüppner H. A heterozygous splice-site mutation in PTHLH causes autosomal dominant shortening of metacarpals and metatarsals. J Bone Miner Res. 2019;34(3):482-489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Albright F, Burnett CH, Smith PH, Parson W. Pseudohypoparathyroidism - an example of “Seabright-Bantam syndrome.” Endocrinology. 1942;30:922-932. [Google Scholar]
- 13. Butcher RW, Sutherland EW. Adenosine 3’,5’-phosphate in biological materials. J Biol Chem. 1962;237(4):1244-1250. [PubMed] [Google Scholar]
- 14. Chase LR, Aurbach GD. Renal adenyl cyclase: anatomically separate sites for parathyroid hormone and vasopressin. Science. 1968;159(3814):545-547. [DOI] [PubMed] [Google Scholar]
- 15. Chase LR, Fedak SA, Aurbach GD. Activation of skeletal adenyl cyclase by parathyroid hormone in vitro. Endocrinology. 1969;84(4):761-768. [DOI] [PubMed] [Google Scholar]
- 16. Chase LR, Aurbach GD. The effect of parathyroid hormone on the concentration of adenosine 3’,5’-monophosphate in skeletal tissue in vitro. J Biol Chem. 1970;245(7):1520-1526. [PubMed] [Google Scholar]
- 17. Chase LR, Aurbach GD. Parathyroid function and the renal excretion of 3’5’-adenylic acid. Proc Natl Acad Sci U S A. 1967;58(2):518-525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chase LR, Melson GL, Aurbach GD. Pseudohypoparathyroidism: defective excretion of 3’,5’-AMP in response to parathyroid hormone. J Clin Invest. 1969;48(10):1832-1844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Farfel Z, Brickman AS, Kaslow HR, Brothers VM, Bourne HR. Defect of receptor-cyclase coupling protein in pseudohypoparathyroidism. N Engl J Med. 1980;303(5):237-242. [DOI] [PubMed] [Google Scholar]
- 20. Levine MA, Downs RW Jr, Singer M, Marx SJ, Aurbach GD, Spiegel AM. Deficient activity of guanine nucleotide regulatory protein in erythrocytes from patients with pseudohypoparathyroidism. Biochem Biophys Res Commun. 1980;94(4):1319-1324. [DOI] [PubMed] [Google Scholar]
- 21. Albright F, Forbes AP, Henneman PH. Pseudo-pseudohypoparathyroidism. Trans Assoc Am Physicians. 1952;65:337-350. [PubMed] [Google Scholar]
- 22. Bray P, Carter A, Simons C, et al. Human cDNA clones for four species of G alpha s signal transduction protein. Proc Natl Acad Sci U S A. 1986;83(23):8893-8897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kozasa T, Itoh H, Tsukamoto T, Kaziro Y. Isolation and characterization of the human Gsa gene. Proc Natl Acad Sci USA. 1988;85:2081-2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Patten JL, Johns DR, Valle D, et al. Mutation in the gene encoding the stimulatory G protein of adenylate cyclase in Albright’s hereditary osteodystrophy. N Engl J Med. 1990;322(20):1412-1419. [DOI] [PubMed] [Google Scholar]
- 25. Weinstein LS, Gejman PV, Friedman E, et al. Mutations of the Gs a-subunit gene in Albright hereditary osteodystrophy detected by denaturing gradient gel electrophoresis. Proc Natl Acad Sci USA. 1990;87:8287-8290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Davies SJ, Hughes HE. Imprinting in Albright’s hereditary osteodystrophy. J Med Genet. 1993;30(2):101-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lemos MC, Thakker RV. GNAS mutations in Pseudohypoparathyroidism type 1a and related disorders. Hum Mutat. 2015;36(1):11-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Drezner M, Neelon FA, Lebovitz HE. Pseudohypoparathyroidism type II: a possible defect in the reception of the cyclic AMP signal. N Engl J Med. 1973;289(20):1056-1060. [DOI] [PubMed] [Google Scholar]
- 29. O’Donnell JK, Sweet RW, Stadel JM. Expression and characterization of the long and short splice variants of GS alpha in S49 cyc- cells. Mol Pharmacol. 1991;39(6):702-710. [PubMed] [Google Scholar]
- 30. Cooper DM, Boyajian CL, Goldsmith PK, Unson CG, Spiegel A. Differential expression of low molecular weight form of Gs-alpha in neostriatum and cerebellum: correlation with expression of calmodulin-independent adenylyl cyclase. Brain Res. 1990;523(1):143-146. [DOI] [PubMed] [Google Scholar]
- 31. Thiele S, Werner R, Ahrens W, et al. A disruptive mutation in exon 3 of the GNAS gene with Albright hereditary osteodystrophy, normocalcemic pseudohypoparathyroidism, and selective long transcript variant Gsalpha-L deficiency. J Clin Endocrinol Metab. 2007;92(5):1764-1768. [DOI] [PubMed] [Google Scholar]
- 32. Puzhko S, Goodyer CG, Kerachian MA, et al. Parathyroid hormone signaling via Gαs is selectively inhibited by an NH(2)-terminally truncated Gαs: implications for pseudohypoparathyroidism. J Bone Miner Res. 2011;26(10):2473-2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hayward BE, Moran V, Strain L, Bonthron DT. Bidirectional imprinting of a single gene: GNAS1 encodes maternally, paternally, and biallelically derived proteins. Proc Natl Acad Sci U S A. 1998;95(26):15475-15480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ischia R, Lovisetti-Scamihorn P, Hogue-Angeletti R, Wolkersdorfer M, Winkler H, Fischer-Colbrie R. Molecular cloning and characterization of NESP55, a novel chromogranin-like precursor of a peptide with 5-HT1B receptor antagonist activity. J Biol Chem. 1997;272(17):11657-11662. [DOI] [PubMed] [Google Scholar]
- 35. Hayward BE, Bonthron DT. An imprinted antisense transcript at the human GNAS1 locus. Hum Mol Genet. 2000;9(5):835-841. [DOI] [PubMed] [Google Scholar]
- 36. Abramowitz J, Grenet D, Birnbaumer M, Torres HN, Birnbaumer L. XLalphas, the extra-long form of the alpha-subunit of the Gs G protein, is significantly longer than suspected, and so is its companion Alex. Proc Natl Acad Sci U S A. 2004;101(22):8366-8371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hayward BE, Barlier A, Korbonits M, et al. Imprinting of the G(s)alpha gene GNAS1 in the pathogenesis of acromegaly. J Clin Invest. 2001;107(6):R31-R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Mantovani G, Ballare E, Giammona E, Beck-Peccoz P, Spada A. The gsalpha gene: predominant maternal origin of transcription in human thyroid gland and gonads. J Clin Endocrinol Metab. 2002;87(10):4736-4740. [DOI] [PubMed] [Google Scholar]
- 39. Germain-Lee EL, Ding CL, Deng Z, et al. Paternal imprinting of Galpha(s) in the human thyroid as the basis of TSH resistance in pseudohypoparathyroidism type 1a. Biochem Biophys Res Commun. 2002;296(1):67-72. [DOI] [PubMed] [Google Scholar]
- 40. Liu J, Erlichman B, Weinstein LS. The stimulatory G protein alpha-subunit Gs alpha is imprinted in human thyroid glands: implications for thyroid function in pseudohypoparathyroidism types 1A and 1B. J Clin Endocrinol Metab. 2003;88(9):4336-4341. [DOI] [PubMed] [Google Scholar]
- 41. Weinstein LS, Xie T, Qasem A, Wang J, Chen M. The role of GNAS and other imprinted genes in the development of obesity. Int J Obes (Lond). 2010;34(1):6-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Weinstein LS, Yu S, Ecelbarger CA. Variable imprinting of the heterotrimeric G protein G(s) alpha-subunit within different segments of the nephron. Am J Physiol Renal Physiol. 2000;278(4):F507-F514. [DOI] [PubMed] [Google Scholar]
- 43. Tafaj O, Hann S, Ayturk U, Warman ML, Jüppner H. Mice maintain predominantly maternal Gαs expression throughout life in brown fat tissue (BAT), but not other tissues. Bone. 2017;103(10):177-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Usardi A, Mamoune A, Nattes E, Carel JC, Rothenbuhler A, Linglart A. Progressive development of PTH resistance in patients with inactivating mutations on the maternal allele of GNAS. J Clin Endocrinol Metab. 2017;102(6):1844-1850. [DOI] [PubMed] [Google Scholar]
- 45. Linglart A, Gensure RC, Olney RC, Jüppner H, Bastepe M. A novel STX16 deletion in autosomal dominant pseudohypoparathyroidism type Ib redefines the boundaries of a cis-acting imprinting control element of GNAS. Am J Hum Genet. 2005;76(5):804-814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Turan S, Fernandez-Rebollo E, Aydin C, et al. Postnatal establishment of allelic Gαs silencing as a plausible explanation for delayed onset of parathyroid hormone resistance owing to heterozygous Gαs disruption. J Bone Miner Res. 2014;29(3):749-760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Yu S, Yu D, Lee E, et al. Variable and tissue-specific hormone resistance in heterotrimeric Gs protein alpha-subunit (Gsalpha) knockout mice is due to tissue-specific imprinting of the gsalpha gene. Proc Natl Acad Sci U S A. 1998;95(15):8715-8720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Gejman PV, Weinstein LS, Martinez M, et al. Genetic mapping of the Gs-alpha subunit gene (GNAS1) to the distal long arm of chromosome 20 using a polymorphism detected by denaturing gradient gel electrophoresis. Genomics. 1991;9(4):782-783. [DOI] [PubMed] [Google Scholar]
- 49. Peters J, Wroe SF, Wells CA, et al. A cluster of oppositely imprinted transcripts at the Gnas locus in the distal imprinting region of mouse chromosome 2. Proc Natl Acad Sci U S A. 1999;96(7):3830-3835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Vlaeminck-Guillem V, D’herbomez M, Pigny P, et al. Pseudohypoparathyroidism Ia and hypercalcitoninemia. J Clin Endocrinol Metab. 2001;86(7):3091-3096. [DOI] [PubMed] [Google Scholar]
- 51. Mantovani G, Maghnie M, Weber G, et al. Growth hormone-releasing hormone resistance in pseudohypoparathyroidism type Ia: new evidence for imprinting of the Gs alpha gene. J Clin Endocrinol Metab. 2003;88(9):4070-4074. [DOI] [PubMed] [Google Scholar]
- 52. Grüters-Kieslich A, Reyes M, Sharma A, et al. Early-onset obesity: unrecognized first evidence for GNAS mutations and methylation changes. J Clin Endocrinol Metab. 2017;102(8):2670-2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Shoemaker AH, Jüppner H. Nonclassic features of pseudohypoparathyroidism type 1A. Curr Opin Endocrinol Diabetes Obes. 2017;24(1):33-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Linglart A, Carel JC, Garabédian M, Lé T, Mallet E, Kottler ML. GNAS1 lesions in pseudohypoparathyroidism Ia and Ic: genotype phenotype relationship and evidence of the maternal transmission of the hormonal resistance. J Clin Endocrinol Metab. 2002;87(1):189-197. [DOI] [PubMed] [Google Scholar]
- 55. Fernández-Rebollo E, Lecumberri B, Gaztambide S, Martinez-Indart L, Perez de Nanclares G, Castaño L; Spanish PHP Group . Endocrine profile and phenotype-(epi)genotype correlation in Spanish patients with pseudohypoparathyroidism. J Clin Endocrinol Metab. 2013;98(5):E996-1006. [DOI] [PubMed] [Google Scholar]
- 56. Elli FM, deSanctis L, Ceoloni B, et al. Pseudohypoparathyroidism type Ia and pseudo-pseudohypoparathyroidism: the growing spectrum of GNAS inactivating mutations. Hum Mutat. 2013;34(3):411-416. [DOI] [PubMed] [Google Scholar]
- 57. Thiele S, Werner R, Grötzinger J, et al. A positive genotype-phenotype correlation in a large cohort of patients with Pseudohypoparathyroidism Type Ia and Pseudo-pseudohypoparathyroidism and 33 newly identified mutations in the GNAS gene. Mol Genet Genomic Med. 2015;3(2):111-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Thiele S, Mantovani G, Barlier A, et al. From pseudohypoparathyroidism to inactivating PTH/PTHrP signalling disorder (iPPSD), a novel classification proposed by the EuroPHP network. Eur J Endocrinol. 2016;175(6):P1-P17. [DOI] [PubMed] [Google Scholar]
- 59. Pereda A, Elli FM, Thiele S, et al. Inactivating PTH/PTHrP signaling disorders (iPPSDs): evaluation of the new classification in a multicenter large series of 544 molecularly characterized patients. Eur J Endocrinol. 2021;184(2):311-320. [DOI] [PubMed] [Google Scholar]
- 60. Long DN, McGuire S, Levine MA, Weinstein LS, Germain-Lee EL. Body mass index differences in pseudohypoparathyroidism type 1a versus pseudopseudohypoparathyroidism may implicate paternal imprinting of Galpha(s) in the development of human obesity. J Clin Endocrinol Metab. 2007;92(3):1073-1079. [DOI] [PubMed] [Google Scholar]
- 61. Mouallem M, Shaharabany M, Weintrob N, et al. Cognitive impairment is prevalent in pseudohypoparathyroidism type Ia, but not in pseudopseudohypoparathyroidism: possible cerebral imprinting of Gsalpha. Clin Endocrinol (Oxf). 2008;68(2):233-239. [DOI] [PubMed] [Google Scholar]
- 62. Hanna P, Grybek V, Perez de Nanclares G, et al. Genetic and epigenetic defects at the GNAS locus lead to distinct patterns of skeletal growth but similar early-onset obesity. J Bone Miner Res. 2018;33(8):1480-1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Turan S, Thiele S, Tafaj O, et al. Evidence of hormone resistance in a pseudo-pseudohypoparathyroidism patient with a novel paternal mutation in GNAS. Bone. 2015;71(2):53-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Geneviève D, Sanlaville D, Faivre L, et al. Paternal deletion of the GNAS imprinted locus (including Gnasxl) in two girls presenting with severe pre- and post-natal growth retardation and intractable feeding difficulties. Eur J Hum Genet. 2005;13(9):1033-1039. [DOI] [PubMed] [Google Scholar]
- 65. Richard N, Molin A, Coudray N, Rault-Guillaume P, Jüppner H, Kottler ML. Paternal GNAS mutations lead to severe intrauterine growth retardation (IUGR) and provide evidence for a role of XLαs in fetal development. J Clin Endocrinol Metab. 2013;98(9):E1549-E1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Plagge A, Gordon E, Dean W, et al. The imprinted signaling protein XL alpha s is required for postnatal adaptation to feeding. Nat Genet. 2004;36(8):818-826. [DOI] [PubMed] [Google Scholar]
- 67. Chudoba I, Franke Y, Senger G, et al. Maternal UPD 20 in a hyperactive child with severe growth retardation. Eur J Hum Genet. 1999;7(5):533-540. [DOI] [PubMed] [Google Scholar]
- 68. Kawashima S, Nakamura A, Inoue T, et al. Maternal uniparental disomy for chromosome 20: physical and endocrinological characteristics of five patients. J Clin Endocrinol Metab. 2018;103(6):2083-2088. [DOI] [PubMed] [Google Scholar]
- 69. Mulchandani S, Bhoj EJ, Luo M, et al. Maternal uniparental disomy of chromosome 20: a novel imprinting disorder of growth failure. Genet Med. 2016;18(4):309-315. [DOI] [PubMed] [Google Scholar]
- 70. Ahmed SF, Barr DG, Bonthron DT. GNAS1 mutations and progressive osseous heteroplasia. N Engl J Med. 2002;346(21):1669-1671. [DOI] [PubMed] [Google Scholar]
- 71. Adegbite NS, Xu M, Kaplan FS, Shore EM, Pignolo RJ. Diagnostic and mutational spectrum of progressive osseous heteroplasia (POH) and other forms of GNAS-based heterotopic ossification. Am J Med Genet A. 2008;146A(14):1788-1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Pignolo RJ, Ramaswamy G, Fong JT, Shore EM, Kaplan FS. Progressive osseous heteroplasia: diagnosis, treatment, and prognosis. Appl Clin Genet. 2015;8:37-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Liu J, Litman D, Rosenberg MJ, Yu S, Biesecker LG, Weinstein LS. A GNAS1 imprinting defect in pseudohypoparathyroidism type IB. J Clin Invest. 2000;106(9):1167-1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Weinstein LS, Yu S, Warner DR, Liu J. Endocrine manifestations of stimulatory G protein alpha-subunit mutations and the role of genomic imprinting. Endocr Rev. 2001;22(5):675-705. [DOI] [PubMed] [Google Scholar]
- 75. Levine MA. Pseudohypoparathyroidism. In: Bilezikian JP, Raisz LG, Rodan GA. ed. Principles of Bone Biology. Academic Press; 2002:1137-1159. [Google Scholar]
- 76. Mantovani G, Bondioni S, Linglart A, et al. Genetic analysis and evaluation of resistance to thyrotropin and growth hormone-releasing hormone in pseudohypoparathyroidism type Ib. J Clin Endocrinol Metab. 2007;92(9):3738-3742. [DOI] [PubMed] [Google Scholar]
- 77. Pérez de Nanclares G, Fernández-Rebollo E, Santin I, et al. Epigenetic defects of GNAS in patients with pseudohypoparathyroidism and mild features of Albright’s hereditary osteodystrophy. J Clin Endocrinol Metab. 2007;92(6):2370-2373. [DOI] [PubMed] [Google Scholar]
- 78. Mariot V, Maupetit-Méhouas S, Sinding C, Kottler ML, Linglart A. A maternal epimutation of GNAS leads to Albright osteodystrophy and parathyroid hormone resistance. J Clin Endocrinol Metab. 2008;93(3):661-665. [DOI] [PubMed] [Google Scholar]
- 79. Unluturk U, Harmanci A, Babaoglu M, et al. Molecular diagnosis and clinical characterization of pseudohypoparathyroidism type-Ib in a patient with mild Albright’s hereditary osteodystrophy-like features, epileptic seizures, and defective renal handling of uric acid. Am J Med Sci. 2008;336(1):84-90. [DOI] [PubMed] [Google Scholar]
- 80. Mantovani G, de Sanctis L, Barbieri AM, et al. Pseudohypoparathyroidism and GNAS epigenetic defects: clinical evaluation of Albright hereditary osteodystrophy and molecular analysis in 40 patients. J Clin Endocrinol Metab. 2010;95(2):651-658. [DOI] [PubMed] [Google Scholar]
- 81. Sanchez J, Perera E, Jan de Beur S, et al. Madelung-like deformity in pseudohypoparathyroidism type 1b. J Clin Endocrinol Metab. 2011;96(9):E1507-E1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Sharma A, Phillips AJ, Jüppner H. Hypoplastic Metatarsals–Beyond Cosmesis. N Engl J Med. 2015;373(22):2189-2190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Zazo C, Thiele S, Martín C, et al. ; Spanish PHP Group . Gsα activity is reduced in erythrocyte membranes of patients with pseudohypoparathyroidism due to epigenetic alterations at the GNAS locus. J Bone Miner Res. 2011;26(8):1864-1870. [DOI] [PubMed] [Google Scholar]
- 84. Bréhin AC, Colson C, Maupetit-Méhouas S, et al. Loss of methylation at GNAS exon A/B is associated with increased intrauterine growth. J Clin Endocrinol Metab. 2015;100(4):E623-E631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Winter JS, Hughes IA. Familial pseudohypoparathyroidism without somatic anomalies. Can Med Assoc J. 1980;123(1):26-31. [PMC free article] [PubMed] [Google Scholar]
- 86. Jüppner H, Schipani E, Bastepe M, et al. The gene responsible for pseudohypoparathyroidism type Ib is paternally imprinted and maps in four unrelated kindreds to chromosome 20q13.3. Proc Natl Acad Sci U S A. 1998;95(20):11798-11803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Bastepe M, Pincus JE, Sugimoto T, et al. Positional dissociation between the genetic mutation responsible for pseudohypoparathyroidism type Ib and the associated methylation defect at exon A/B: evidence for a long-range regulatory element within the imprinted GNAS1 locus. Hum Mol Genet. 2001;10(12):1231-1241. [DOI] [PubMed] [Google Scholar]
- 88. Bastepe M, Fröhlich LF, Hendy GN, et al. Autosomal dominant pseudohypoparathyroidism type Ib is associated with a heterozygous microdeletion that likely disrupts a putative imprinting control element of GNAS. J Clin Invest. 2003;112(8):1255-1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Elli FM, de Sanctis L, Peverelli E, et al. Autosomal dominant pseudohypoparathyroidism type Ib: a novel inherited deletion ablating STX16 causes loss of imprinting at the A/B DMR. J Clin Endocrinol Metab. 2014;99(4):E724-E728. [DOI] [PubMed] [Google Scholar]
- 90. Yang Y, Chu X, Nie M, et al. A novel long-range deletion spanning STX16 and NPEPL1 causing imprinting defects of the GNAS locus discovered in a patient with autosomal-dominant pseudohypoparathyroidism type 1B. Endocrine. 2020;69(1):212-219. [DOI] [PubMed] [Google Scholar]
- 91. Fröhlich LF, Bastepe M, Ozturk D, Abu-Zahra H, Jüppner H. Lack of Gnas epigenetic changes and pseudohypoparathyroidism type Ib in mice with targeted disruption of syntaxin-16. Endocrinology. 2007;148(6):2925-2935. [DOI] [PubMed] [Google Scholar]
- 92. Richard N, Abeguilé G, Coudray N, et al. A new deletion ablating NESP55 causes loss of maternal imprint of A/B GNAS and autosomal dominant pseudohypoparathyroidism type Ib. J Clin Endocrinol Metab. 2012;97(5):E863-E867. [DOI] [PubMed] [Google Scholar]
- 93. Perez-Nanclares G, Velayos T, Vela A, Muñoz-Torres M, Castaño L. Pseudohypoparathyroidism type Ib associated with novel duplications in the GNAS locus. PLoS One. 2015;10(2):e0117691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Nakamura A, Hamaguchi E, Horikawa R, et al. Complex genomic rearrangement within the GNAS region associated with familial pseudohypoparathyroidism type 1b. J Clin Endocrinol Metab. 2016;101(7):2623-2627. [DOI] [PubMed] [Google Scholar]
- 95. Reyes M, Kagami M, Schnabel D, Fukami M, Jüppner H. A novel GNAS duplication associated with loss-of-methylation restricted to exon A/B causes pseudohypoparathyroidism type Ib (PHP1B). J Bone Miner Res. Published online November 12, 2020. doi: 10.1002/jbmr.4209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Grigelioniene G, Nevalainen PI, Reyes M, et al. A large inversion involving GNAS Exon A/B and all exons encoding Gsα is associated with autosomal dominant pseudohypoparathyroidism type Ib (PHP1B). J Bone Miner Res. 2017;32(4):776-783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Bastepe M, Fröhlich LF, Linglart A, et al. Deletion of the NESP55 differentially methylated region causes loss of maternal GNAS imprints and pseudohypoparathyroidism type Ib. Nat Genet. 2005;37(1):25-27. [DOI] [PubMed] [Google Scholar]
- 98. Takatani R, Molinaro A, Grigelioniene G, et al. Analysis of multiple families with single individuals affected by pseudohypoparathyroidism type Ib (PHP1B) reveals only one novel maternally inherited GNAS deletion. J Bone Miner Res. 2016;31(4):796-805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Chillambhi S, Turan S, Hwang DY, Chen HC, Jüppner H, Bastepe M. Deletion of the noncoding GNAS antisense transcript causes pseudohypoparathyroidism type Ib and biparental defects of GNAS methylation in cis. J Clin Endocrinol Metab. 2010;95(8):3993-4002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Fröhlich LF, Mrakovcic M, Steinborn R, Chung UI, Bastepe M, Jüppner H. Targeted deletion of the Nesp55 DMR defines another Gnas imprinting control region and provides a mouse model of autosomal dominant PHP-Ib. Proc Natl Acad Sci U S A. 2010;107(20):9275-9280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Liu J, Nealon JG, Weinstein LS. Distinct patterns of abnormal GNAS imprinting in familial and sporadic pseudohypoparathyroidism type IB. Hum Mol Genet. 2005;14(1):95-102. [DOI] [PubMed] [Google Scholar]
- 102. Linglart A, Bastepe M, Jüppner H. Similar clinical and laboratory findings in patients with symptomatic autosomal dominant and sporadic pseudohypoparathyroidism type Ib despite different epigenetic changes at the GNAS locus. Clin Endocrinol (Oxf). 2007;67(6):822-831. [DOI] [PubMed] [Google Scholar]
- 103. Fernández-Rebollo E, Pérez de Nanclares G, Lecumberri B, et al. Exclusion of the GNAS locus in PHP-Ib patients with broad GNAS methylation changes: evidence for an autosomal recessive form of PHP-Ib? J Bone Miner Res. 2011;26(8):1854-1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Elli FM, Linglart A, Garin I, et al. The prevalence of GNAS deficiency-related diseases in a large cohort of patients characterized by the EuroPHP network. J Clin Endocrinol Metab. 2016;101(10):3657-3668. [DOI] [PubMed] [Google Scholar]
- 105. Elli FM, de Sanctis L, Bollati V, et al. Quantitative analysis of methylation defects and correlation with clinical characteristics in patients with pseudohypoparathyroidism type I and GNAS epigenetic alterations. J Clin Endocrinol Metab. 2014;99(3):E508-E517. [DOI] [PubMed] [Google Scholar]
- 106. Jan de Beur S, Ding C, Germain-Lee E, Cho J, Maret A, Levine M. Discordance between genetic and epigenetic defects in pseudohypoparathyroidism type 1b revealed by inconsistent loss of maternal imprinting at GNAS1. Am J Hum Genet. 2003;73:314-322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Maupetit-Méhouas S, Mariot V, Reynès C, et al. Quantification of the methylation at the GNAS locus identifies subtypes of sporadic pseudohypoparathyroidism type Ib. J Med Genet. 2011;48(1):55-63. [DOI] [PubMed] [Google Scholar]
- 108. Bastepe M, Lane AH, Jüppner H. Paternal uniparental isodisomy of chromosome 20q–and the resulting changes in GNAS1 methylation–as a plausible cause of pseudohypoparathyroidism. Am J Hum Genet. 2001;68(5): 1283-1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Bastepe M, Altug-Teber O, Agarwal C, Oberfield SE, Bonin M, Jüppner H. Paternal uniparental isodisomy of the entire chromosome 20 as a molecular cause of pseudohypoparathyroidism type Ib (PHP-Ib). Bone. 2011;48(3):659-662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Dixit A, Chandler KE, Lever M, et al. Pseudohypoparathyroidism type 1b due to paternal uniparental disomy of chromosome 20q. J Clin Endocrinol Metab. 2013;98(1):E103-E108. [DOI] [PubMed] [Google Scholar]
- 111. Takatani R, Minagawa M, Molinaro A, et al. Similar frequency of paternal uniparental disomy involving chromosome 20q (patUPD20q) in Japanese and Caucasian patients affected by sporadic pseudohypoparathyroidism type Ib (sporPHP1B). Bone. 2015;79(10):15-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Colson C, Decamp M, Gruchy N, et al. High frequency of paternal iso or heterodisomy at chromosome 20 associated with sporadic pseudohypoparathyroidism 1B. Bone. 2019;123(6):145-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Gahurova L, Tomizawa SI, Smallwood SA, et al. Transcription and chromatin determinants of de novo DNA methylation timing in oocytes. Epigenetics Chromatin. 2017;10:25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Liu J, Yu S, Litman D, Chen W, Weinstein LS. Identification of a methylation imprint mark within the mouse Gnas locus. Mol Cell Biol. 2000;20(16):5808-5817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Williamson CM, Ball ST, Nottingham WT, et al. A cis-acting control region is required exclusively for the tissue-specific imprinting of Gnas. Nat Genet. 2004;36(8):894-899. [DOI] [PubMed] [Google Scholar]
- 116. Mehta S, Williamson CM, Ball S, et al. Transcription driven somatic DNA methylation within the imprinted Gnas cluster. PLoS One. 2015;10(2):e0117378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Xie Y, Wu B, Jin Y, et al. Oocyte-specific deletion of Gsα induces oxidative stress and deteriorates oocyte quality in mice. Exp Cell Res. 2018;370(2):579-590. [DOI] [PubMed] [Google Scholar]
- 118. Snanoudj S, Molin A, Colson C, et al. Maternal transmission ratio distortion of GNAS loss-of-function mutations. J Bone Miner Res. 2020;35(5):913-919. [DOI] [PubMed] [Google Scholar]
- 119. Kiuchi Z, Reyes M, Jüppner H. Preferential maternal transmission of STX16-GNAS mutations responsible for autosomal dominant pseudohypoparathyroidism type Ib (PHP1B): another example of transmission ratio distortion. J Bone Miner Res. Published online November 28, 2020. doi: 10.1002/jbmr.4221 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data sharing is not applicable to this article as no datasets were generated or analyzed during the current study.