Figure 2.
Targeted CRISPR-Cas9 knockout screen implicates SMC5/6 complex localization factor 2 in silencing of unintegrated virus
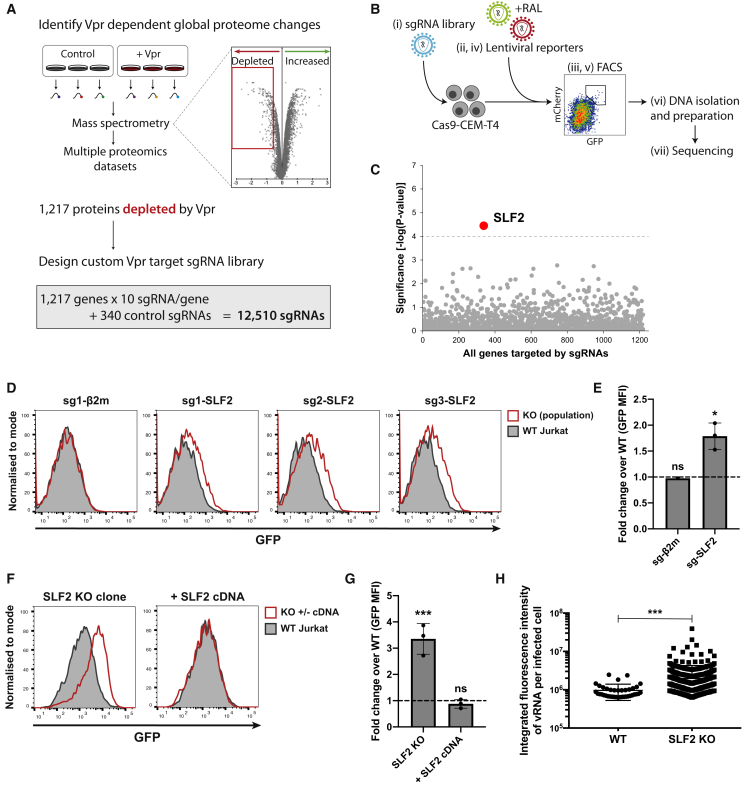
(A–C) CRISPR-Cas9 knockout screen for unintegrated virus silencing factors. (A) A custom sgRNA library was constructed, containing sgRNAs targeting genes encoding Vpr-depleted proteins identified from existing proteomics datasets and used for a CRISPR-Cas9 knockout screen outlined in (B). A pooled Vpr library knockout population (Bi) was infected with GFP and mCherry-lentiviral reporters in presence of RAL (Bii). Rare high expressing cells were enriched by FACS (Biii) followed by repeated reporter virus infection (Biv) and sorting (Bv). DNA was isolated from sorted and unsorted library populations (Bvi) and prepared for next-generation sequencing (Bvii). (C) Candidate genes essential for unintegrated virus silencing were identified using MAGeCK. Genes scoring above multiple-testing-corrected threshold are highlighted.
(D–G) Validation of screen hit. Unintegrated virus reporter infection of mixed knockout (KO) populations 7 days post-sgRNA transduction of Cas9-Jurkat. Flow cytometry 72 hpi (D), quantified as fold change GFP MFI over WT Jurkat (E). Representative example (n = 3). (F) Unintegrated virus reporter infection of clonal SLF2 KO cell line ± full-length SLF2 cDNA complementation, data from n = 3 quantified in (G). (H) Unintegrated Vpr-deletion NL4-3 reporters produce more vRNA upon SLF2 KO. WT or SLF2 KO Jurkat T cells were infected with ΔVpr NL4-3GFP in presence of RAL. 48 hpi, viral RNA was detected by in situ hybridization and quantified as previously described. Data are representative example of n = 2. Error bars show standard deviation. ns, p > 0.05; ∗p < 0.05; ∗∗∗p < 0.001.
See also Figures S2–S4.