Figure 6.
The SMC5/6 complex induces compaction of unintegrated lentiviral genomes
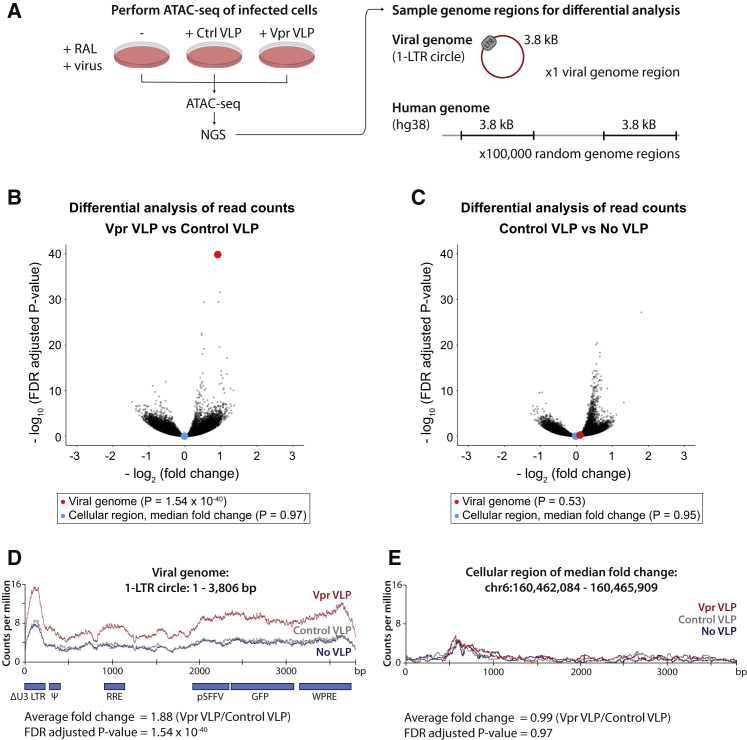
(A–E) ATAC-seq shows increased unintegrated virus chromatin accessibility upon Vpr-mediated SLF2 depletion. (A) WT Jurkat cells were co-infected with a GFP reporter lentivirus and control or Vpr VLPs in the presence of RAL, and ATAC-seq performed 48 hpi. Reads were aligned to the human and viral 1-LTR circle reference genomes. To quantify effects on chromatin accessibility, 100,000 cellular genome regions of equal size to the viral genome (3.8 kB) were randomly defined, and the fold change in normalized sequence coverage between conditions calculated for each region, viral or cellular. For each data point, significance was determined using Fisher’s t test, adjusted by FDR to correct for multiple testing. Comparisons were visualized as volcano plots summarizing data for all 100,000 cellular regions and the viral genome (plotting regions with >50 aligned reads): (B) comparing data for Vpr VLP with control VLP and (C) comparing data for control VLPs with No VLPs. The viral genome (red) and the cellular region of median fold change (blue) are highlighted as enlarged data points for each comparison. Normalized read density for the viral genome is displayed in (D) and for a representative cellular region of median fold change in (E).
See also Figure S7.