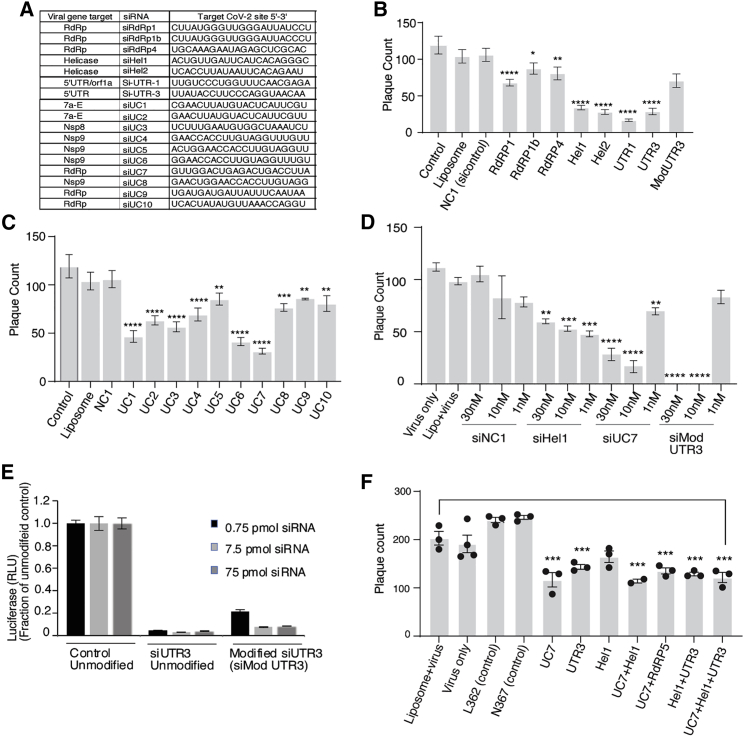
Figure 1.
siRNA screening against SARS-CoV-2
(A) The top candidate siRNAs selected for screening against SARS-CoV-2.16 VeroE6 cells were either pre-treated without (Liposome, Lipo+virus) or with 30 nM of siRNA complexed with Lipofectamine 2000 for 24 h before infection. Viral plaques were counted after 4 days. (B and C) siRNAs targeting genes (B) and phylogenetically conserved regions (C) were tested. (D) The top repressive siRNAs were screened for dose-dependent repression of SARS-CoV-2. (E) The resultant unmodified siRNA controls and the modified siMod UTR3 were transfected with a pSI-Check reporter vector with the 5′ UTR cloned downstream of Renilla luciferase, and knockdown of luciferase activity of the modified siRNA determined relative to the unmodified control. The average of triplicate-treated HEK293 cells is shown with the standard deviation. (F) Combinations of the top candidate siRNAs were selected, mixed in equal molar ratios to a final concentration of 30 nM, and assessed for repression of SARS-CoV-2 in vitro. For (B)–(D) and (F), triplicate treated cells are shown with the standard error of the mean of triplicate treatments and ∗p < 0.05,17 ∗∗p < 0.01, ∗∗∗p < 0.001, and ∗∗∗∗p < 0.001 were considered statistically significant as determined by one-way ANOVA analysis (Dunnett’s post-test) when compared against virus only (control).