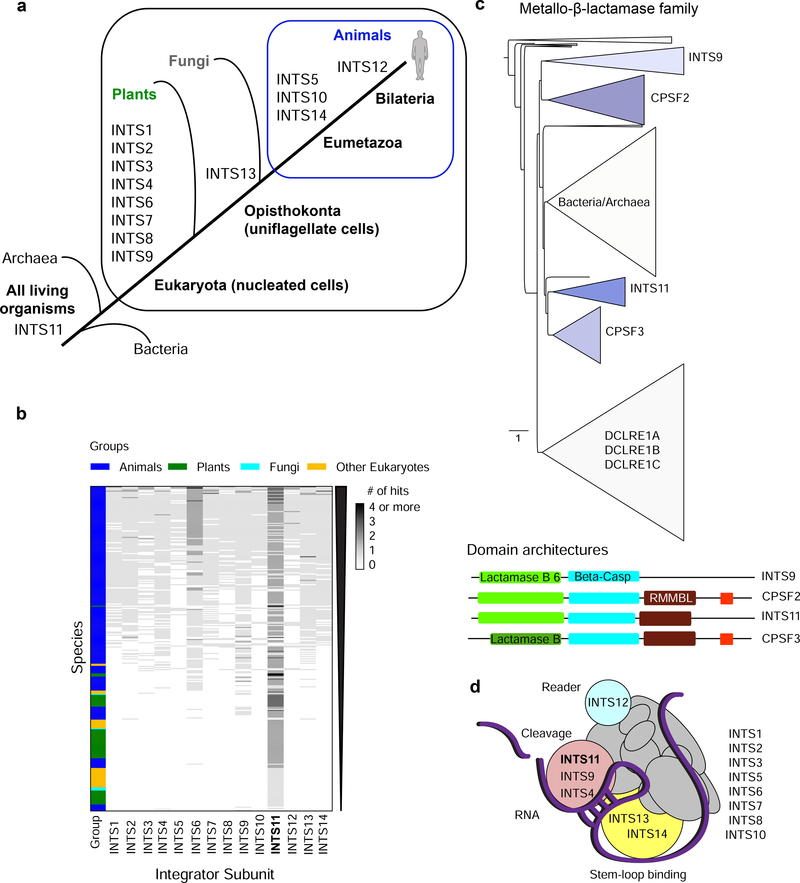
Figure 1. Evolution of the Integrator complex.
a) Overview of the INTS gene ages throughout evolution. Eukarya (nucleated cells), Opisthokonta (uniflagellate cells), Eumetazoa (animals, except sponges), Bilateria (animals with bilateral body symmetry and three germ layers). b) Heatmap of the presence/absence of Integrator-homologous proteins in >300 species (compared to full-length human sequences with the following cut-offs: 30% pair-wise sequence identity, and 50% coverage of the human proteins, in addition to blastp default parameters [29]). Grayscale accounts for the number of hits in one species that aligned to a human INTS. c) Phylogenetic tree of the metallo-β-lactamase superfamily from eggNOG 4.5.1 (COG1236) [33]. Triangle length indicates divergence and triangle height the number of species. Below: domain architecture of human INTS9, CPSF2, INTS11, and CPSF3, as retrieved from Pfam 33.1 [32]. In red CPSF73–100_C domain. d) Model of INTS and their modular interactions. Cleavage module depicted in pink, stem-loop binding module in yellow, and reader module in cyan. Bold: catalytic subunit INTS11.