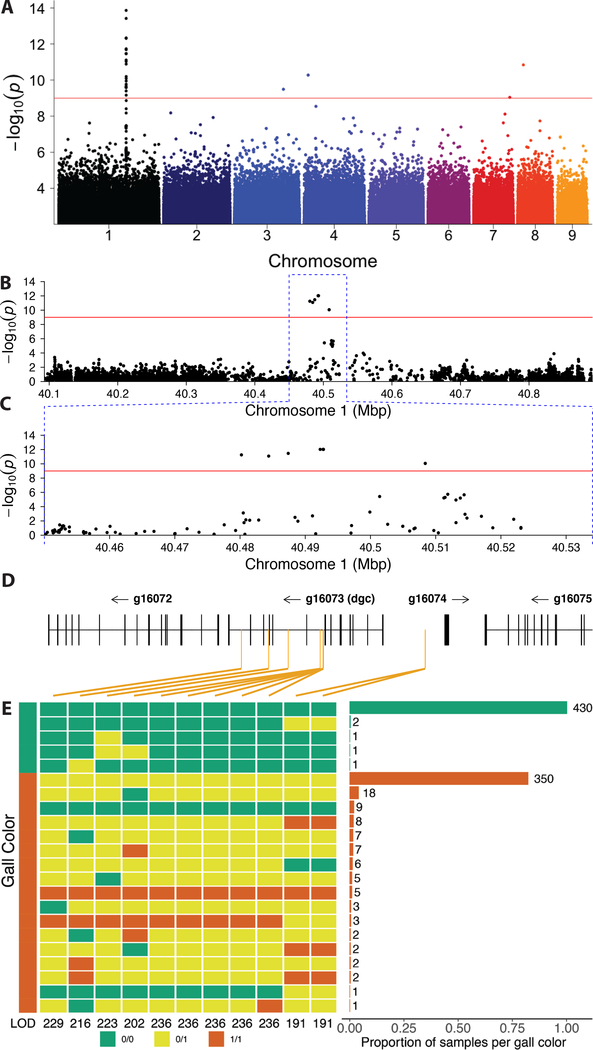
Figure 2. A genome-wide association study (GWAS) identifies variation within a novel aphid gene associated with gall color.
(A) A GWAS of fundatrices isolated from 43 green and 47 red galls identified a small region on chromosome 1 of the H. cornu genome that is strongly associated with gall color. Red line indicates FDR = 0.05. Colors of points on chromosomes are arbitrary.
(B-D) Resequencing 800 kbp of Chromosome 1 centered on the most significant SNPs from the original GWAS to approximately 60X coverage identified 11 spatially clustered SNPs significantly associated with gall color located within the introns and upstream of g16073, which was named dgc (D). (Some SNPs are closely adjacent and cannot be differentiated at this scale.) Significant SNPs are indicated with orange vertical lines in (D).
(E) Genotypes of all 11 SNPs associated with gall color from an independent sample of aphids from 435 green and 431 red galls. Color of gall for aphid samples shown on left and genotype at each SNP is shown adjacent in green (0/0, homozygous ancestral state), yellow (0/1, heterozygous), or red (1/1, homozygous derived state). LOD scores for association with gall color shown for each SNP at bottom of genotype plot. Histogram of frequencies of each multilocus genotype ordered by frequency and collected within gall color is shown on the right. All 11 SNPs are strongly associated with gall color (P < 10−192), and a cluster of 5 SNPs in a 61bp region are most strongly associated with gall color. Individuals homozygous for ancestral alleles at all or most loci and making red galls likely carry variants at other loci that influence gall color (STAR Methods).
See also Figures S1 and S2.