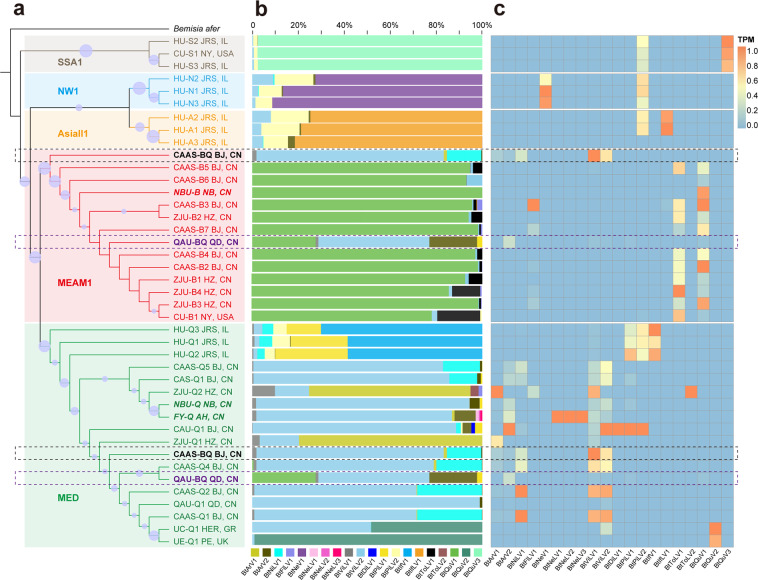
Fig. 3. Correlation between whitefly cryptic species and insect-specific viruses.
a Phylogeny of whitefly cryptic species based on mtCOI sequences using the maximum likelihood method. The mtCOI of Bemisia afer (MK360160.1) was used to root the tree. Two datasets (CAAS-BQ BJ,CN and QAU-BQ QD,CN) containing mixed cryptic species (MEAM1 and MED) are highlighted by dotted frames colored with black and purple, respectively. Nodes with bootstrap values >50% are marked with solid blue circles. b Composition of insect-specific viruses (ISVs) in each whitefly cryptic species dataset. ISVs are color-coded and the names of viruses are indicated at the bottom of the figure. c Relative abundance of ISVs across the different whitefly cryptic species datasets. The transcripts per million (TPM) of each ISV are displayed by the heat map. Abbreviations of the cities and countries: JRS, IL: Jerusalem, Israel; NY: New York, USA; AJ, IN: Ajitgarh, India; BJ, CN: Beijing, China; NB, CN: Ningbo, China; HZ, CN: Hangzhou, China; QD, CN: Qingdao, China; AH, CN: Anhui, China; HER, GR: Heraklion, Greece; PE: Penryn. Abbreviation and details of the whitefly datasets and newly discovered ISVs are listed in Tables 1 and 2, respectively. Abbreviations of the whitefly cryptic species: MEAM1 Middle East Asia Minor 1, MED Mediterranean, NW1 New World 1, SSA1 sub-Saharan Africa 1.