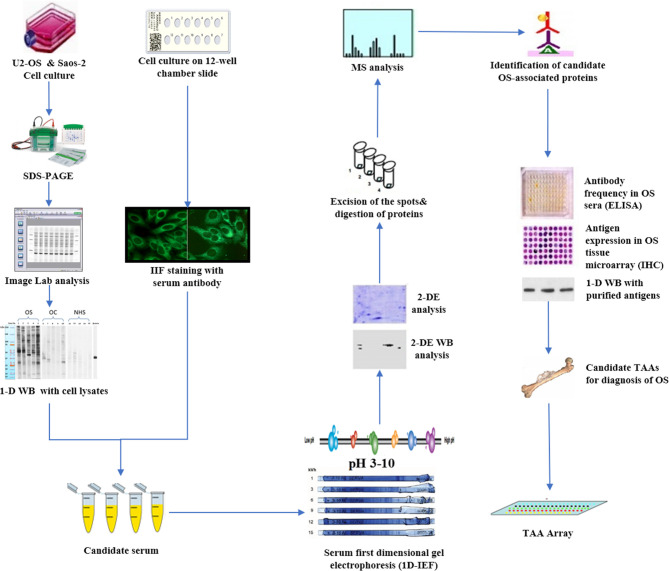
Figure 1.
Schematic representation of identification and validation of TAAs using serological proteome analysis (SERPA) approach. In brief, the sera from OS patients and controls were initially examined using extracts of culture cells as a source of antigens in Western blotting and by indirect immunofluorescence (IIF) on whole cells. With these two techniques, we identify sera which have high-titer fluorescent staining or strong signals to cell extracts on Western blotting and narrow the targeting proteins on specific molecular weight bands, and subsequently use the antibodies in these sera as probes in immunoproteomic screening. Cell extracts of cultured human cells was also applied onto the first-dimension gel (isoelectrofocusing gel), and subsequently loaded onto the second-dimension gel (2-DE-SDS-PAGE). The proteins were transferred to the nitrocellulose membrane or visualized by silver staining or Coomassie brilliant blue staining. After immunoblotting with OS sera and control sera, a number of protein spots of interest were excised from the 2-DE gels, digested by trypsin, and subsequently analyzed by mass spectrometry (MS). In subsequent studies, we will characterize the identified cellular proteins that are potential biomarkers in OS.